†These authors contributed equally.
Academic Editor: Changsoo Kim
Background: Species of the genus Torreya are similar in morphology, and their morphological taxonomic characteristics are not stable because of environmentally induced changes. Therefore, morphology is insufficient for understanding their relationships. Chloroplast genome sequencing technology provides a powerful tool for molecular analysis to get more infomation for classification and identification of Torreya genus. Methods: A total of 4 chloroplast genome of Torreya, including T. Parvifolia, T. nucifera, T. fargesii var. Yunnanensis and T. grandis var. jiulongshanensis, were sequenced and annotated. Campartive genome and phylogenetic tree were provided for variation analysis. Results: The chloroplast genome size of the four samples is about 137 kb, the inverted repeat (IR) regions are identified in the genus Torreya. Genome comparison using mVISTA showed high sequence similarity among different species. Regions with divergence in exon regions include accD, ndhB, ndhF, psbA, psbJ, rpl2, rps3, rps16, rps18, ycf1, and ycf2. The phylogenetic tree based on 73 single-copy genes showed a clearer relationships among different species of Torreya. Conclusions: All genomes of the four Torreya species consist of two short IR regions, and results of the phylogenetic analysis concluded that T. parvifolia should be considered as T. fargesii var. yunnanensis or treated as a sister species. T. grandis var. jiulongshanensis should be treated as a variety of T. grandis according to molecular evidence, supporting the originally published proposal.
The genus Torreya has a deep history, with the earliest fossils of the genus found in Europe being dated to approximately 170 million years ago, within the Jurassic period [2]. Because of the separation of continents, Torreya species were gradually distributed in North America and evolved distinct traits. Then, likely due to the drying up of the Turgai Sea, they migrated into Asia [2, 3]. Because of climate change, human activity, and other factors, trees of this genus exist only in North America and East Asia, showing an obviously disjunct distribution.
The genus Torreya was originally proposed by Arnott in 1838, who published its type species Torreya taxifolia [4]. In 1846, the species of Taxus nucifera was designated by Siebold & Zucc, belonging to the genus Torreya [5]. T. californica was described in 1854 by John Torrey [6] and in 1857, T. grandis was published [7]. In 1899, Franchet published T. fargesii [8] whilst T. jackii was described in 1925 [9]. In 1975, T. yunnanensis was distinguished from T. fargesii in Flora Reipublicae Popularis Sinicae [10]. In 1995, Kang and Tang published T. grandis var. jiulongshanensis according to the morphological characteristics of the leaves. These authors also identified T. fargesii and T. grandis as two different species according to their morphological (especially endosperm) differences, geographic distributions and ecological characteristics. In addition, the authors considered Silba’s change of T. fargesii to be a variety of T. grandis in 1984 as unreasonable and treated T. yunnanensis as a variety of T. fargesii [11]. In 2006, Yi et al. [12] published a new species of the Taxaceae, T. parvifolia. In 2010, Farjon placed T. fargesii as a species in the genus Torreya and treated T. fargesii var. yunnanensis as a variety of T. fargesii, which was consistent with the opinion of Kang and Tang (1995) and the classifications described in the Flora of China [13, 14]. In 2017, based on a comparative analysis of leaf morphology in the genus, Torreya, T. grandis var. jiulongshanensis was treated as an independent species [15]. However, this variety was recently proven to be a natural hybrid species of T. grandis and T. jackii by analysis of gene fragments [16].
In summary, according to the latest classification data, there are currently 7 species and 2 varieties of the genus Torreya worldwide, which are distributed in East Asia and North America and have important economic and scientific value [17, 18, 19]. T. taxifolia and T. californica are native to North America. T. nucifera is widely distributed in Japan, and arbors in the Korean Peninsula were introduced into China and planted as garden trees. There are 4 species and 2 varieties indigenous to China, namely, T. parvifolia, T. fargesii var. yunnanensis, T. grandis var. jiulongshanensis, T. grandis, T. fargesii, and T. jackii. T. clarnensis is an extinct species and was first described from a series of isolated fossil seeds in chert [20]. In this study, in order to determine the characteristics of T. grandis var. jiulongshanensis, chloroplast genomes of T. nucifera and T. fargesii var. yunnanensis were sequenced and compared by our team.
Phylogenetic relationships within the genus Torreya based on the nrDNA-ITS sequence, have strongly indicated that T. fargesii is closely related to T. fargesii var. yunnanensis, and there is no significant difference between T. fargesii and T. nucifera, suggesting that it is better to merge the two species into one [21]. Research using the trnl-trnf sequence supports combining T. fargesii var. yunnanensis into T. fargesii or treating it as a variety of T. fargesii [22]. Research using the psbA-trnH sequence combined with endosperm characteristics supports the treatment of T. fargesii var. yunnanensis as a variety of T. fargesii [23].
Chloroplasts play an important role in plant photosynthesis, moreover, they encode many key proteins important in other metabolic processes [24]. In general, the genome of chloroplasts is typically divided into four regions, including a large single-copy (LSC) region, a small single-copy (SSC) region and two inverted repeat (IRa and IRb) regions [25, 26, 27]. Chloroplast genomes are usually between 120 kb and 170 kb in length [28]. The chloroplast genome is the most valuable reference point for understanding plant evolution and phylogenetic relationships [23] and can be used in molecular phylogenetic and molecular ecological studies because of its highly conserved nature [29]. Phylogenomic analysis based on chloroplast genomes of the genus Torreya in Asia and North America has been performed, illustrating that T. fargesii var. yunnanensis is the sister species of T. nucifera + T. fargesii [1].
At present, the species relationship within the genus Torreya is still unclear. In this study, we aim to use the chloroplast genome to conduct a phylogenetic analysis of genus Torreya in China. We sequenced the chloroplast genomes of T. parvifolia, T. nucifera, T. fargesii var. yunnanensis and T. grandis var. jiulongshanensis. This study focused on the chloroplast genome structure, similarities, and phylogenetic relationships of members of the genus Torreya in East Asia by using maximum likelihood (ML) and Bayesian inference (BI) methods based on single-copy orthologous genes. Clarification of the phylogenetic statuses of these species could help understand their relationships.
Four samples from China were sequenced in this study, and other genomes of the genus Torreya in East Asia were downloaded from GenBank database for comparison analysis (Table 1). Healthy young leaves were collected from each plant, wiped clean, numbered and placed in Ziplock bags. A large amount of color-changing silica gel was used for rapid drying.
The Hi-DNAsecure Plant Kit DP350 (Tiangen Biotech, Beijing, China) was used for DNA extraction following the manufacturer’s instructions (include manufacturer).
| Family | Species | Location | GenBank accession number | SRA accession number |
| Taxaceae | T. parvifolia | Wandun mountain, Wuyi township, Butuo county, Liangshan Yi Autonomous Prefecture, Sichuan province, China | MN244711 | SRR10769481 |
| T. nucifera | Nanjing University, Nanking, Jiangsu province (Jinling University introduced the species from Japan), China | MN244713 | SRR10768423 | |
| T. fargesii var. yunnanensis | Weixi County, Diqing Tibetan Autonomous Prefecture, Yunnan province, China | MN244712 | SRR10758697 | |
| T. grandis var. jiulongshanensis | Jiulong Mountain Nature Reserve, Suichang County, Lishui, Zhejiang province, China | MN244714 | SRR10758782 | |
| T. grandis | Non available | NC_034806 | - | |
| T. fargesii | China | NC_029398 | - | |
| T. jackii | Non available | KX902234 | - | |
| T. jackii | Hangzhou Botanical Garden, China | MK249064 | - | |
| T. nucifera | Lushan Botanical Garden, China | MK249060 | - | |
| T. fargesii var. yunnanensis | Lijiang, Yunnan, China | MK249061 | - | |
| T. californica | Royal Botanic Garden Edinburgh, UK | MK249062 | - | |
| T. taxifolia | Atlanta Botanical Garden, USA | MK249063 | - | |
| Taxus baccata | Non available | NC_035066 | - | |
| Taxus canadensis | Non available | NC_041499 | - | |
| Cephalotaxus oliver | Wuhan Botanical Garden, China | NC_021110 | - | |
| Cephalotaxus sinensis | Taibai Mountain, Shaanxi, China | MF977938 | - | |
| Amentotaxus argotaenia | Wuhan Botanical Garden, China | NC_027581 | - | |
| Amentotaxus formosana | Academia Sinica and Taipei Botanical Garden, Taiwan, China | NC_024945 | - | |
| Podocarpaceae | Podocarpus lambertii | Lages, Santa Catarina, Brazil | NC_023805 | - |
| SRA, sequence read archive. | ||||
PE150 paired-end sequencing was conducted on an Illumina HiSeq X Ten genomic
sequencer (Illumina Inc., San Diego, California, USA) at Majorbio Company
(Shanghai, China). All sequencing depths were over 100
MITObim v1.8 [30] was used to assemble the chloroplast genome with default parameters, and the final circular structure was formed manually. Annotations of these chloroplast genomes were performed by using online software GeSeq [31] and BLAST+ 2.9.0 [32] with an E-value of 1e-5. The genome map was illustrated by using OGDRAW [33]. Annotated chloroplast genome sequences were submitted to GenBank and raw data were uploaded to Sequence Read Archive (SRA) of NCBI and will be released upon publication. GenBank accession numbers of genomes used in this study are listed in Table 1.
Seven chloroplast genomes of the genus Torreya, including the four new genomes sequenced in this work, were compared by using mVISTA [34, 35] based on the Shuffle-LAGAN method [36].
The seven chloroplast genomes of Torreya species were also compared with BLAST Ring Image Generator (BRIG) v0.95 [37], and BLAST+ v2.9.0 was used with an E-value of 1e-5.
Single-copy orthologous genes were selected from the results analyzed by OrthoFinder v2.2.7 [38, 39]. We used MAFFT v7.45 for multiple sequence alignment with the L-INS-i strategy for more accuracy [40, 41]. Then, JModelTest v2.1.1.0 [42, 43] was used to find the best-fitting models for single-copy orthologous genes according to the Akaike information criterion (AIC), where the lowest value showed the best fit [44].
A phylogenetic tree based on ML with single-copy orthologous genes was built by
using RAxML v8.2.4 [45] with the best-fitting model and 1000 bootstrap
replicates. A BI-based phylogenetic tree was constructed with single-copy
orthologous genes in MrBayes v3.2.7 [46, 47, 48], running for 2
Results for the four sequenced genomes, including genome structure and GC content, are shown in Table 2. The genome sizes of T. parvifolia, T. nucifera, T. fargesii var. yunnanensis, and T. grandis var. jiulongshanensis are 136781 bp, 136955 bp, 136807 bp, and 137320 bp, with an average genome GC content of 35.49%, 35.46%, 35.49%, and 35.41%, respectively. Their large single-copy (LSC) regions are 97386 bp, 97516 bp, 97421 bp, and 114484 bp, respectively. Their small single-copy (SSC) regions are 38799 bp, 38841 bp, 38790 bp, and 22148 bp, respectively. The inverted repeat (IR) regions are 298 bp, 299 bp, 298 bp, and 344 bp, respectively. All of the genomes have similar small IR regions.
| Statistics | T. parvifolia | T. nucifera | T. fargesii var. yunnanensis | T. grandis var. jiulongshanensis |
| Genome size (bp) | 136781 | 136955 | 136807 | 137320 |
| LSC (bp) | 97386 | 97516 | 97421 | 114484 |
| SSC (bp) | 38799 | 38841 | 38790 | 22148 |
| IRa (bp) | 298 | 299 | 298 | 344 |
| IRb (bp) | 298 | 299 | 298 | 344 |
| Total genome GC (%) | 35.49 | 35.46 | 35.49 | 35.41 |
| LSC GC (%) | 35.24 | 35.22 | 35.24 | 35.57 |
| SSC GC (%) | 36.15 | 36.1 | 36.15 | 34.45 |
| IR GC (%) | 33.67 | 33.56 | 33.33 | 41.11 |
| LSC, large single-copy region; SSC, small single-copy region; IR, inverted repeat region. | ||||
Chloroplast genome maps of T. parvifolia, T. nucifera, T. fargesii var. yunnanensis, and T. grandis var. jiulongshanensis are shown in Fig. 1. T. parvifolia, T. nucifera, and T. fargesii var. yunnanensis are very similar in gene number, order and names. However, T. grandis var. jiulongshanensis has a different gene number, order and name, especially gene order. In contrast to the other three taxa, T. grandis var. jiulongshanensis lacks the rps11 gene but has the clpP gene as a unique gene. The locations of some genes in the chloroplast genome maps differ from others. For example, in T. grandis var. jiulongshanensis, the atpA, atpF, atpH, atpI, rpoB, rpoC1, rpoC2, rps2, rps4, psaA, psaB, psbC and psbD genes are located in the LSC region, and the ndhF and ycf1 genes are located in the SSC region. However, in the other three species, the former group are located in the SSC region, and the latter group are located in the LSC region.
 Fig. 1.
Fig. 1.Chloroplast genome maps of four species of the genus Torreya in East Asia. (A) The chloroplast genome map of T. parvifolia. Genes outside the circle transcribe counterclockwise, while those inside transcribe clockwise. Genes with different functions are marked with different colors. (B) The chloroplast genome map of T. nucifera. Genes outside the circle transcribe counterclockwise, while those inside transcribe clockwise. Genes with different functions are marked with different colors. (C) The chloroplast genome map of T. fargesii var. yunnanensis. Genes outside the circle transcribe counterclockwise, while those inside transcribe clockwise. Genes with different functions are marked with different colors. (D) The chloroplast genome map of T. grandis var. jiulongshanensis. Genes outside the circle transcribe counterclockwise, while those inside transcribe clockwise. Genes with different functions are marked with different colors.
Seven chloroplast genomes of species in the genus Torreya, compared by mVISTA with the Shuffle-LAGAN method and annotated according to T. jackii MK249064, are shown in Fig. 2. Overall, the sequence similarity is high among these species, especially in exon regions. Regions with such divergence in exon regions include accD, ndhB, ndhF, psbA, psbJ, rpl2, rps3, rps16, rps18, ycf1, and ycf2. tRNA or rRNA genes with high levels of divergence include trnT-GGU, trnR-UCU, trnF-GAA, trnL-CAA, trnQ-UUG, trnN-GUU, trnM-CAU, rrn4.5 and rrn5. A comparison of the chloroplast genomes of seven species of the genus Torreya at the level of whole genome sequence was analyzed using BRIG, providing information for identifying the unique sequence of the chloroplast genome (Fig. 3). The sequence of T. jackii MK249064 was selected as a reference.
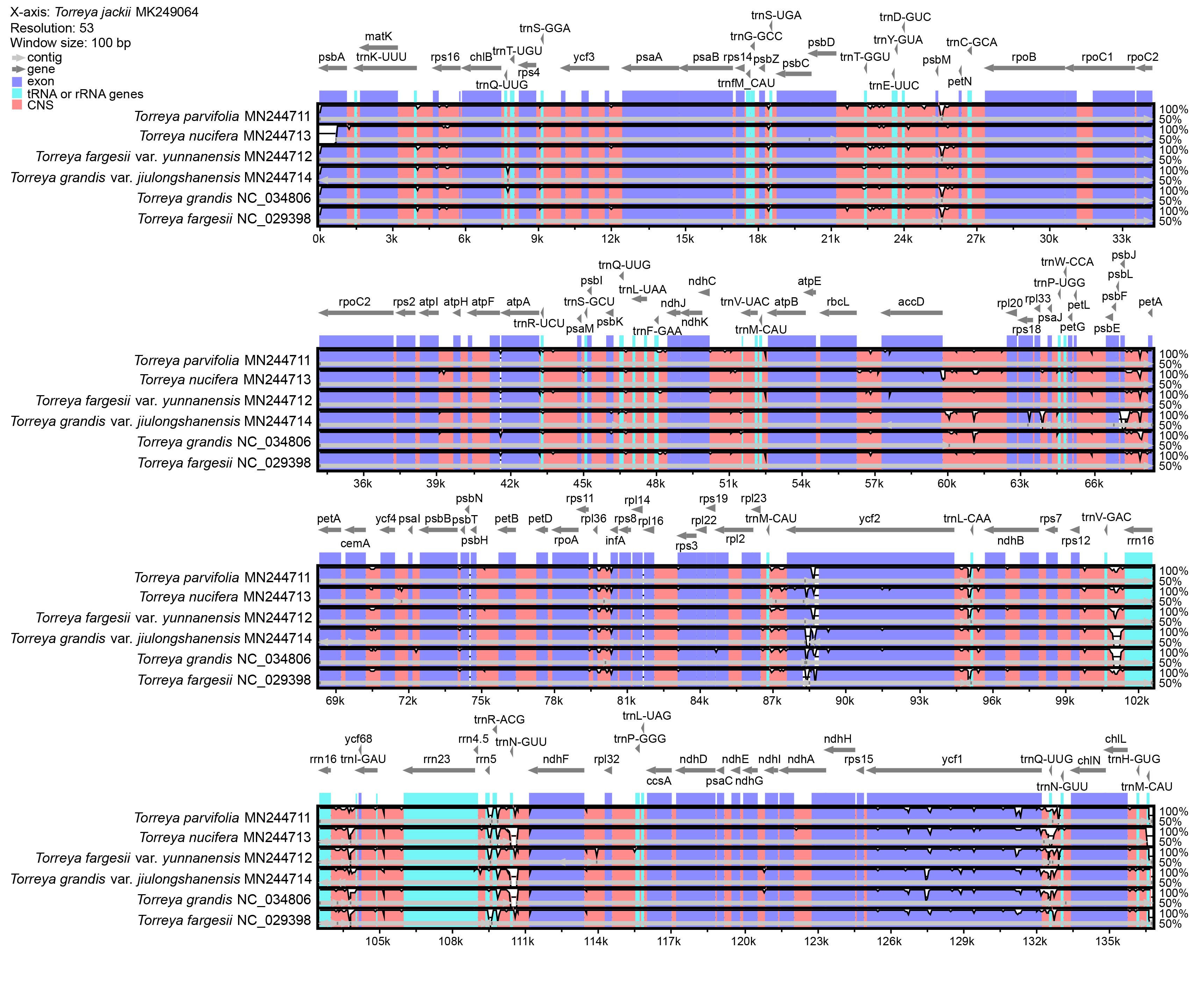 Fig. 2.
Fig. 2.Comparison of chloroplast genomes of seven species of the genus Torreya by using mVISTA based on Shuffle-LAGAN method. The sequence of T. jackii MK249064 is selected as reference. The thick gray arrow at the top of the array indicates gene orientations. The dark-blue regions, light-blue regions, and pink regions represent exon, tRNA or rRNA genes, and conserved non-coding sequences (CNS), respectively.
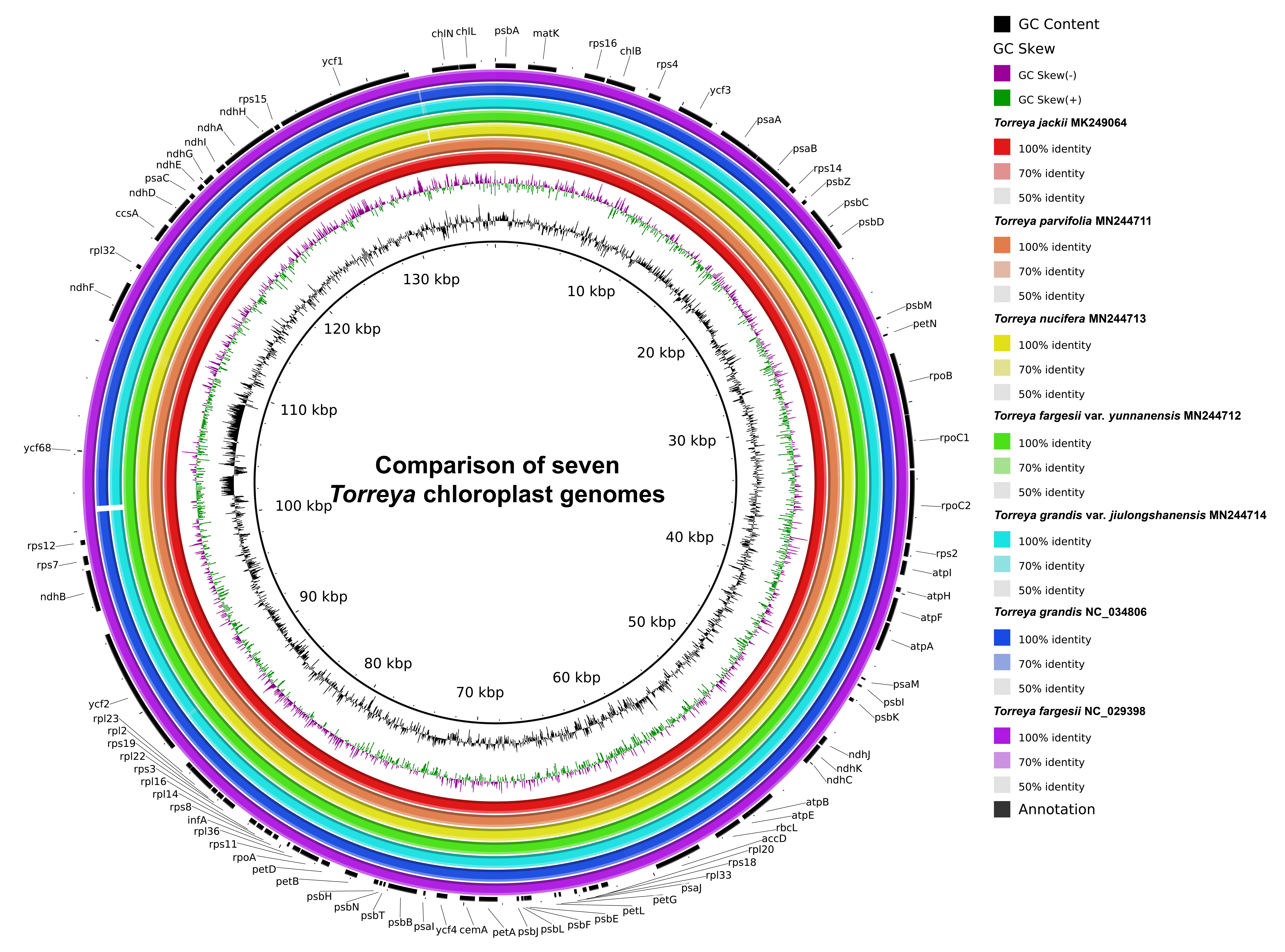 Fig. 3.
Fig. 3.Comparison of chloroplast genomes of seven species of the genus Torreya by using BRIG. The sequence of T. jackii MK249064 is selected as reference. Two of the innermost rings show GC content and GC skew from inside to outside, respectively.
Seventy-three single-copy orthologous genes were selected (Supplementary Table 1), and then a phylogenetic tree was built based on the ML and BI methods with these genes using the best-fitting model GTR + GAMMA + I. The ML and BI phylogenetic trees show the same relationships among the Torreya species (Fig. 4 ). T. parvifolia MN244711 is located closely to T. fargesii var. yunnanensis MN244712, and they are included in a clade with T. fargesii var. yunnanensis MK249061. T. fargesii NC_029398 and T. nucifera MK249060 form a single clade. T. parvifolia MN244711 and T. fargesii var. yunnanensis MN244712 are close to the clade containing T. fargesii NC_029398 and T. nucifera MK249060. T. grandis var. jiulongshanensis MN244714 is close to T. californica MK249062, and this clade is close to T. grandis NC_034806. The clade including T. grandis var. jiulongshanensis MN244714, T. californica MK249062 and T. grandis NC_034806 is close to our sample of T. nucifera MN244713. T. jackii KX902234 and T. jackii MK249064 are located in the most exterior part of the phylogenetic tree. The genus Torreya forms one clade in the phylogenetic tree. Based on phylogenetic trees, T. nucifera MK249060 is close to T. fargesii NC_029398, different from the results of Zhang et al. [1].
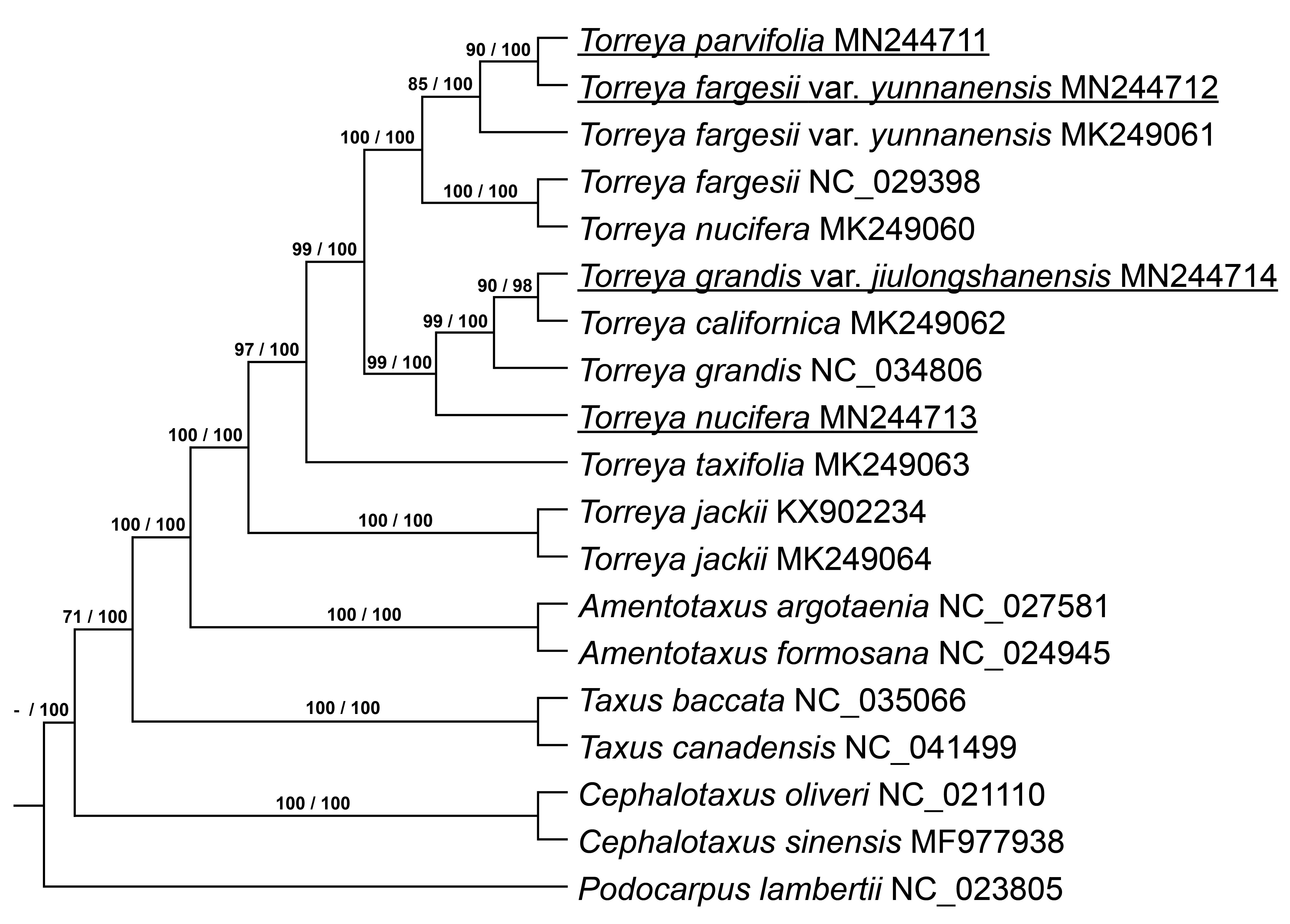 Fig. 4.
Fig. 4.Phylogenetic tree shows relationship and supported values. Both ML and BI tree show the same relationship. Supported values of ML and BI tree are showed on branch and divide by sign of division, respectively.
Statistics of the genomes of four Torreya species showed that the IR regions were very short, with only 298–344 bp. Shrinkage or loss of the IR region is not rare in plants, and in a study of Pinus thunbergii, the IR region was found to have been reduced to 495 bp [50]. Loss of the IR region occurred in all cases of Pisum sativum L., Vicia faba L. [51], Glyptostrobus pensilis [52] and Cryptomeria japonica [53]. Pinaceae and non-Pinaceae conifers independently lost different copies of IRs, according to comparisons of the junctions near LSC regions and residual IR copies among gymnosperms [54], while the lack of an IR copy was considered a derived characteristic common to all conifers and “a single loss event defining the conifers as a monophyletic group” [55]. In addition, the four species shared similar GC contents (35.41%–35.49%). T. parvifolia, T. nucifera, and T. fargesii var. yunnanensis showed very similar chloroplast genome map structures, while T. grandis var. jiulongshanensis exhibited a very different order of genes. However, according to comparisons by both mVISTA and BRIG, there were no great differences among these species, especially in protein-coding regions. Therefore, this specificity could be due to genetic recombination.
In a 2006 publication, T. parvifolia was treated as a new species [13]. However, based on the phylogenetic analysis reported here, T. parvifolia MN244711 and T. fargesii var. yunnanensis MN244712 have a close relationship, with a high correlation and form one clade with T. fargesii var. yunnanensis MK249061. Therefore, we believe that the species T. parvifolia should be considered as T. fargesii var. yunnanensis or treated as a sister species according to the molecular evidence. T. nucifera MK249060 was collected from Lushan Botanical Garden, China, and the material of T. nucifera MN244713 used in this study was collected from Nanjing University, Nanking, Jiangsu Province (Jinling University introduced the species from Japan), which has been verified by historical evidence. In the recent study [1], T. nucifera MK249060 and T. fargesii NC_029398 formed a single cluster with a very high association based on the ML method, and this cluster was closest to T. fargesii var. yunnanensis MK249061, followed by T. grandis NC_034806. However, in this study, T. nucifera MN244713 was closest to T. grandis NC_034806 and T. grandis var. jiulongshanensis MN244714 and then to T. fargesii NC_029398 and T. fargesii var. yunnanensis MK249061. Therefore, the molecular biological evidence in this study supports the treatment of T. nucifera MK249060 as T. fargesii.
In a recent study by Kou et al. [15], phylogenetic analysis of the nuclear internal transcribed spacer (ITS) and combined sequences of chloroplast rbcL and rpl16 genes in the genus Torreya showed that T. grandis var. jiulongshanensis was close to T. jackii according to the combination sequences based on the ML method. However, it is insufficient evidence to suggest that T. grandis var. jiulongshanensis is a natural hybrid of T. jackii and T. grandis, basing on a few sequence fragments. In addition, the study comparing leaf variation among T. grandis var. jiulongshanensis, T. grandis, T. fargesii and T. jackii [16] proposed that T. grandis var. jiulongshanensis be treated as an independent species rather than a variety of T. grandis due to the morphological features of their leaves. The two studies above, examining gene fragments or just leaf variations, failed to offer hard evidence for related taxonomy. In our study, phylogenetic trees based on the whole genome-wide level single-copy orthologous genes showed consistent results by ML method and BI method, both illustrating that T. grandis var. jiulongshanensis MN244714 was close to T. grandis NC_034806. Therefore, according to our study, T. grandis var. jiulongshanensis should be treated as a variety of T. grandis rather than a natural hybrid between T. jackii and T. grandis or an independent species, supporting the original proposal [12].
In this study, we analyzed the complete cp genomes of four Torreya species. These genomes provided a basic genetic tool for species identification within the genus. All genomes sequenced by us, two short IR regions were identified in the work. We compared the cp genomes of different species of Torreya, and results showed that the gene size, content, and order were all similar. In contrast to the other three taxa, T. grandis var. jiulongshanensis lacks the rps11 gene but has the clpP gene as a unique gene. Results of the phylogenetic analysis concluded that T. parvifolia should be considered as T. fargesii var. yunnanensis or treated as a sister species. T. grandis var. jiulongshanensis should be treated as a variety of T. grandis according to molecular evidence, supporting the originally published proposal.Generally, this study provides valuable genetic information of Torreya which can aid in further phylogenetic studies, species identification, and evolutionary relationships.
SRA, Sequence read archive; BRIG, BLAST Ring Image Generator; AIC, Akaike Information Criterion; ML, maximum likelihood; BI, Bayesian inference; LSC, large single-copy; SSC, small single-copy; IR, Inverted repeat.
ZPM, XNN, LH and XH performed the experiments and data analysis. RBW and JHL collected samples. ZPM and XH wrote the manuscript. BBM and JHL revised the manuscript. ZPM, XNN, contributed equally to this work. All authors have read and approved the final manuscript.
Not applicable.
The authors thank Majorbio company (Shanghai, China) for Genome Sequencing and Winnerbio company (Shanghai, China) for bioinformatic support.
This work was supported by Anhui Province Natural Science Foundation (No. 1908085QC126), Natural Science Foundation of China (No. 31400321), and Opening Project of Zhejiang Provincial Key Laboratory of Plant Evolutionary Ecology and Conservation (No. EEC2014-01).
The authors declare no conflict of interest.
