Academic Editor: Masoud Foroutan
Background: Coronavirus disease-2019 (COVID-19) has become a pandemic around the globe due to the Severe Acute Respiratory Syndrome Corona Virus-2 (SARS-CoV-2), a new variant of the Coronavirus (CoV) family. The rapid transmission of the infectious disease, 135,646,617 positive cases from which 2,930,732 mortality cases were recorded until 11 April 2021. In an emergency, several existing anti-viral, anti-malarial, and anti-HIV drugs have been used on a repurposing basis. However, without proper clinical evidence, it may create several side effects for the patient. Thus, recommending potential and less-toxic regimens at this emergency stage is the most crucial aspect for any physician. Methods: We have hypothesized a combinatorial drug approach against COVID-19 and to select potential combinations from ten anti-HIV drugs and ten vitamin C derivatives were systematically validated using advanced bioinformatic tools. Initially, the chemical structures used as ligands from PubChem and the target protein, SARS-CoV-2 main protease (PDB ID: 6Y84) from the protein data bank were retrieved for this study. Further, assess the potency, toxicity, drug-ability, and pharmacokinetics profiles using several bioinformatics tools, viz., molecular docking by the AutoDock 4.1 software with predicting activity spectra for substances, Molsoft, ProTox, and SwissADME tools. Molecular dynamics simulation was also employed for most potential candidates to assess their binding stability using GROMACS 5.1.4 software. Results: The above computational investigation indicated that ‘darunavir with L-ascorbyl-2,6-dibutyrate or ascorbic acid-2-sulfate’ combinations strongly inhibit the SARS-CoV-2-main protease as a potential treatment option against COVID-19. Mostly, vitamin C derivatives enhanced the anti-COVID activity and might reduce the post-treatment side effects of darunavir in combination. Conclusions: Overall, the present work suggests that bioinformatics tools are suitable for recognizing potential candidates in an emergency, and herein the selected ‘anti-HIV-drug-vitamin c derivatives’ cocktails may potential-cum-fewer toxic regimens against COVID-19 treatment.
The emerging Coronavirus disease-2019 (COVID-19), caused by the Severe Acute
Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), has continuously
added a higher number of morbidity and mortality cases, globally [1, 2, 3].
Under this unrestrained grievous health endemic situation, the World Health
Organization (WHO) has declared it as a Public Health Emergency of International
Concern [1, 4]. The outbreak and quick transmission of the COVID-19 is terrible
than the previously emerged SARS-CoV and any other viral diseases [2, 5]. The
symptoms exhibited by the patients under COVID-19 infection are similar to that
of the previously SARS-CoVs infection, including fever-cough, complicity during
the breath, muscle pain, sore throat and sputum production. However, severe
pneumonia and multi-organ dysfunction under COVID-19 infection have been found to
enhance the mortality rate, predominantly in immune-compromised patients, i.e.,
in the elderly age group (
The food and drug administration (FDA) has not recommended any drug(s) rather
re-purposed a few medications to treat this harmful infectious disease to tackle
the pandemic in an emergency. However, fighting against the emerging aggressive
positive-sense single-strand RNA or (+) ssRNA viral infection without effective
medicine is the most significant panic situation for protecting the global health
system [7, 8]. Indeed, experts or physicians used several alternative
combinations of existing anti-viral, anti-malarial, anti-HIV (human
immunodeficiency virus), and anti-inflammatory drugs on a non-random trial basis
without much clinical experience to control the situation [6, 9, 10]. Mainly,
approved anti-viral drugs such as baloxavir marboxil, darunavir, favipiravir,
lopinavir, oseltamivir, remdesivir, ritonavir, etc., along with several
immune-modulatory, anti-inflammatory remedies such as Fingolimod, Sarilumab and
Tocilizumab are used in multiple combinations against COVID-19 [9, 10, 11]. Even the
combined treatment with obsolete anti-malarial quinine derivatives such as
hydroxyl-chloroquine with the macrolide (azithromycin) and tetracycline
(doxycycline) class of antibiotics is continuously used as an alternative
against COVID-19 [11, 12]. The COVID-19 is an aggregated form of previously
recognized SARS and the Middle East respiratory syndrome (MERS) virus and its
genome was found to have
In parallel, some claims for using certain Traditional Chinese Medicines (TMC) to control COVID-19 [15, 16]. In collaboration with the State Administration of Traditional Chinese Medicine, the Hubei Hospital of Traditional Chinese Medicine filed a trial application to start the phase-I clinical trial with a health-boosting TMC formulation against COVID-19 [17]. Similarly, few also claim for the use of Ayurvedic and Homeopathic formulations from the Indian Unani system and other Western traditional medicines for the treatment of COVID-19 [18, 19]. As per previous reports, homeopathic remedies also successfully prevented Cholera, Spanish Influenza, Yellow fever, and Typhoid diseases [20, 21, 22]. Thus, locating an active and non-toxic/less toxic combinatorial approach is essential in this emergency.
Therefore, the present work also proposed an alternative health-boosting and immune-modulating ‘mainstream-natural combined formula’ against COVID-19. High throughput screening approaches were employed in selecting a specific active anti-HIV drug and a vitamin C derivative (VCD) for their possible practice in combination/synergistic for the treatment of COVID-19.
Based on previous literatures and research findings, ten anti-HIV protease
inhibitor drugs namely, amprenavir (PubChem ID: 65016), ASC09 (PubChem ID:
53361968), atazanavir (PubChem ID: 148192), darunavir (PubChem ID:
213039), indinavir (PubChem ID: 5362440), lopinavir (PubChem ID: 92727),
nelfinavir (PubChem ID: 64143), ritonavir (PubChem ID: 392622), saquinavir
(PubChem ID: 441243), tipranavir (PubChem ID: 54682461) and vitamin Cor L-ascorbic acid (PubChem ID: 54670067) as a code, VCD-1
with another nine VCDs, VCD-2
(2-O-
Initially, each ligand chemical structure was converted to dot PDB (.pdb) data format, as a required format according to the used docking software, AutoDock 4.1. Later on, based on the binding pattern or molecular interactions from high-throughput molecular docking study against SARS-CoV-2 protease, the lead anti-HIV drugs and VCDs were selected [23, 24]. The molecular docking was performed, followed by a multi-step AutoDock tutorial. Again, a double docking study was conducted to assess the combinatorial action of both leading anti-HIV drugs and ascorbic acid derivatives against the same target SARS-CoV-2 protease [25, 26, 27]. The molecular interaction and binding pattern of protein-ligand docking complexes were visualized by BIOVIA DSV v 4.5 software [28].
Furthermore, the physicochemical properties such as molecular weight (MW), number of hydrogen acceptors (H-BA) and donors (H-BD), topological surface area (tPSA), partition or distribution coefficient (XLogP) value, number of rotatable-bonds (RB), molar refractivity (MR), collectively known as standardized Lipinski rules of five (RO5) parameters were recorded from PubChem database. The overall drug-likeness score of individual anti-HIV drugs and VCDs properties was recorded using the tool, MolSoft (https://www.molsoft.com/).
Again, to predict the possible therapeutic potencies such as vaso-protective, respiratory analeptic, immuno-stimulant, antioxidant, anti-inflammatory and anti-viral properties of ascorbic acid and their derivatives through the advanced bioinformatics tool, the Perdition of Activity Spectra for Substances (PASS) (http://www.pharmaexpert.ru/passonline/) was used. The specific biological activities of each candidate as per the present objectives, probable activity (Pa) and probable inactivity (Pi) values were recorded through data mining processes against the training data set presented in the PASS data library.
The tool, ProTox (http://tox.charite.de/protox_II/), was employed to predict
the probable toxicity profiles such as hepatotoxicity, carcinogenicity,
immunotoxicity toxicity, mutagenicity, cytotoxicity, toxicity class and lethal
dose (LD
To check the stability/flexibility of docking complexes, selected two docking
complexes (based on the effective docking score), from each side (one from
anti-HIV drugs, i.e., ‘main protease-darunavir’ and one from VCDs, namely,
‘main-protease- ascorbic acid-2-sulfate or VCD-6’ were selected for MD simulation
with the software GROMACS 5.1.4 (Groningen Machine for Chemical Simulations) in
the GROMOS force field in 50 ns time scale [25, 26, 27]. In the first step, we have
generated topologies files for each complex using the PRODRG server
(http://prodrg1.dyndns.org/submit.html). Selected docking complexes with the
SPC-E water-cubic box model (whose volume was about 976.40, 976.50, 977.48,
977.48 nm
As per the research hypothesis, the docking scores (kcal/mol) were used as a standard parameter for selecting the most efficient combination of any vitamin C derivatives with a potent anti-HIV drug against deadly SARS-CoV-2 (Tables 1,2). The docking score of L-ascorbic acid and derivatives was within -7.00 kcal/mol. Among all derivatives, the VCD-6 (ascorbic acid 2-sulfate) with docking score -6.75 (Fig. 1), and the VCD-3 (L-ascorbyl 2,6-dibutyrate) with docking score -6.19 kcal/mol (Fig. 2) were chosen for further studies (Table 1). On the other hand, all the selected ten anti-HIV drugs had docking scores within -11.00 kcal/mol (Table 2). Darunavir was chosen from all anti-viral drugs as the most active drug against SARS-CoV-2 protease, based on the docking a score, -10.25 kcal/mol (Fig. 3). Therefore, based on the overall docking score and molecular interactions, the VCD-6 and VCD-3 among vitamin C derivatives, and darunavir among all ten anti-viral drugs were selected for further studies.
| VCDs | Docking score in kcal/mol (type of interaction and their bond length) | Drug likeness score | Lethal dose (mg/kg) | Bioavailability score |
| VCD-1 | −4.94 (Lys5: Conventional-H-bond 2.50 Å; Trp207: Conventional-H-bond 2.04 Å; Leu282: Conventional-H-bonds 1.97 Å & 2.19 Å; Ser284: Conventional-H-bond 2.03 Å; Glu288: Conventional-H-bonds 1.81 Å & 2.23 Å) | 0.74 | 3367 | 0.56 |
| VCD-2 | −2.87 (Pro52: Conventional-H-bonds 1.86 & 1.90 Å; Asn53: Conventional-H-bond 1.97 Å; Tyr54: Conventional-H-bond 2.0 Å; Glu55: Conventional-H-bonds 1.92, 2.68 Å) | 0.53 | 25000 | 0.11 |
| VCD-3 | −6.19 (Lys5: Conventional-H-bonds 2.03 Å, 2.31 Å & 2.50 Å; Gly127: Conventional-H-bonds 2.27 Å & 2.91 Å) | 0.34 | 25000 | 0.56 |
| VCD-4 | −4.56 (Arg4: Conventional-H-bond 2.13 Å-Carbon-H-bond 3.03 Å; Lys5: Conventional-H-bond 2.32 Å; Trp207: Conventional-H-bond 2.07 Å; Leu282: Conventional-H-bond 2.20 Å; Ser284: Conventional-H-bonds 1.90, 2.09 & 2.30 Å) | 0.80 | 5000 | 0.56 |
| VCD-5 | −4.78 (Phe3: Carbon-H-bond 3.46 Å; Lys5: Conventional-H-bond 2.0 Å; Trp207: Conventional-H-bond 2.39 Å; Leu282: Conventional-H-bonds 1.72 Å & 1.89 Å; Ser284: Conventional-H-bond 2.11 Å; Glu288: Conventional-H-bond 2.07 Å) | 0.27 | 5000 | 0.56 |
| VCD-6 | −6.75 (Phe3: Conventional-H-bond 2.18 Å; Arg4: Conventional-H-bond 2.10 Å; Lys5: Conventional-H-bonds-1.68, 1.70, 2.18, 2.83, 3.10 Å; Glu288: Conventional-H-bond 2.24 Å) | 0.24 | 10000 | 0.11 |
| VCD-7 | −5.61 (Phe3: Conventional-H-bond-2.17 Å; Arg4: Conventional-H-bond-2.95 Å- Carbon-H-bond 3.70 Å; Trp207: Conventional-H-bond 2.15 Å; Leu282: Conventional-H-bonds 1.81 Å & 2.25 Å; Ser284: Conventional-H-bond 2.34 Å) | 0.11 | 5000 | 0.56 |
| VCD-8 | −5.98 (Lys5: conventional-H-bond 2.05, 2.16, 2.56 & 2.61 Å; Gln127: Conventional-H-bonds 2.19, 2.23 & 2.45 Å) | 0.12 | 5000 | 0.56 |
| VCD-9 | −4.72 (Phe3: Carbon-H-bond 3.71 Å; Lys5: Conventional-H-bonds 2.55, 2.53, 2.58 & 2.71 Å; Leu282: Conventional-H-bonds 1.78 & 2.18 Å; Ser284: Conventional-H-bond 1.78 Å-Carbon-H-bond 3.63 Å) | 0.45 | 5000 | 0.56 |
| VCD-10 | −5.79 (Lys5: Conventional-H-bonds 1.90, 1.93, 2.42 & 2.57 Å; Van125: Conventional-H-bond 2.09 Å; Gln127: Conventional-H-bonds 2.03 & 2.36 Å) | 0.41 | 5000 | 0.56 |
| VCD-1, L-ascorbic acid; VCD-2,
2-O- | ||||
| Anti-HIV drugs | Docking score in kcal/mol (type of interaction and their bond length) | Drug likeness score | Lethal dose (kg/mg) | Bioavailability score |
| Amprenavir | −8.43 (Lys5: Pi-alkyl 5.1 Å-Pi-cation 4.1 Å; Ala7: Pi-alkyl-3.98 Å; Gln127: Conventional-H-bond 1.66 Å-Carbon-H-bond 3.35 Å; Glu290: Pi-anion 3.50 Å) | 1.14 | 300 | 0.55 |
| ASC09 | −9.56 (Lys5: Conventional-H-bonds 1.96 & 2.01 Å-Pi-alkyls 4.76 & 4.82 Å; Ala7: Conventional-H-bond 1.86 Å-Pi-alkyls 4.45 Å & 4.76 Å; Ala116: Pi-alkyl 5.17 Å; Gly124: Pi-sigma-3.84 Å; Val125: Conventional-H-bond 1.72 Å-Carbon-H-bond 2.87 Å; Tyr126: Pi-alkyl 5.01 Å-Pi-sigma 3.52 Å) | 1.09 | 150 | 0.17 |
| Atazanavir | −7.37 (Arg4: Conventional-H-bond-2.42 Å; Lys5: Conventional-H-bond 2.28 Å; Ala7: Pi-alkyl 5.30 Å; Gln127: Carbon-H-bonds 3.28 & 3.44 Å; Lys137: Pi-alkyl 5.30 Å-Pi-cation 4.70 Å; Asp289: Pi-anion 3.69 Å) | 0.11 | 200 | 0.17 |
| Darunavir | −10.25 (Arg4: Pi-alkyl 4.85 Å-Pi-loan pair 2.75 Å; Lys5: Conventional-H-bonds 2.70 & 2.63 Å-Pi-loan pair 3.34 Å-Pi-alkyl 4.43 Å; Tyr126: Pi-alkyls 4.34 & 4.43 Å; Leu282: Conventional-H-bond 2.85 Å; Glu288: Pi-anions 4.17 & 4.20 Å) | 0.60 | 245 | 0.55 |
| Indinavir | −7.80 (Arg4: Conventional-H-bond 2.14 Å-Carbon-H-bond 3.23 Å; Lys137: Pi-alkyls 3.97 & 5.10 Å; Leu286: Pi-alkyl 5.42 Å; Glu288: Pi-anions 3.44 & 3.99 Å) | 1.86 | 5000 | 0.55 |
| Lopinavir | −9.00 (Arg4: Pi-alkyls 5.31 & 5.48 Å; Lys5: Conventional-H-bond-2.20 & 2.66 Å; sulfurs 2.88 & 2.94 Å; Ser284: Conventional-H-bond 2.14 Å; Glu288: Conventional-H-bond-2.07 Å) | 1.10 | 5000 | 0.55 |
| Nelfinavir | −9.97 (Asn133: Conventional H-bond 1.73 Å; Gly195: Conventional H-bond 1.94 Å; Thr198: Pi-sigma 3.67 Å) | 1.41 | 600 | 0.55 |
| Ritonavir | −8.75 (Arg4: Conventional-H-bonds 1.71 & 2.0 Å-Carbon-H-bond 3.76; Lys5: Conventional-H-bonds 2.51 & 2.56 Å; Lys137: Pi-alkyl 4.43 Å; Val171: Pi-alkyl 5.38 Å; Leu286: Pi-alkyl 4.93 Å; Glu288: Pi-alkyl 4.93 Å-Pi-anion 3.41 Å) | 0.11 | 1000 | 0.17 |
| Saquinavir | −8.97 (Lys5: Pi-alkyl 4.48 Å; Ala7: Pi-alkyl 4.10 Å; Gln127: conventional-H-bond 2.44 & 3.04 Å; Lys137: Conventional-H-bond 2.23 Å; Leu286: Pi-alkyl 4.64 & 5.11 Å) | 0.69 | 500 | 0.17 |
| Tipranavir | −9.98 (Lys5: Pi-alkyl 4.62 Å-Pi-cation 4.64 Å; Ala7: Pi-alkyl 4.46 Å; Tyr126: Pi-alkyl 3.93 Å; Gln127: Conventional-H-bond 1.77 Å; Cys128: Pi-alkyl 4.76 Å; Glu288: Pi-anion 3.13 & 3.44 Å) | 0.72 | 333 | 0.56 |
 Fig. 1.
Fig. 1.Molecular interaction of most effective VCD, VCD-6 (ascorbic acid 2-sulfate) with docking score -6.75 kcal/mol against SARS-CoV-2 protease from docking study.
 Fig. 2.
Fig. 2.Molecular interaction of most effective vitamin C derivative, VCD-3 (L-ascorbyl 2,6-dibutyrate) with docking score -6.19 kcal/mol against SARS-CoV-2 protease from docking study.
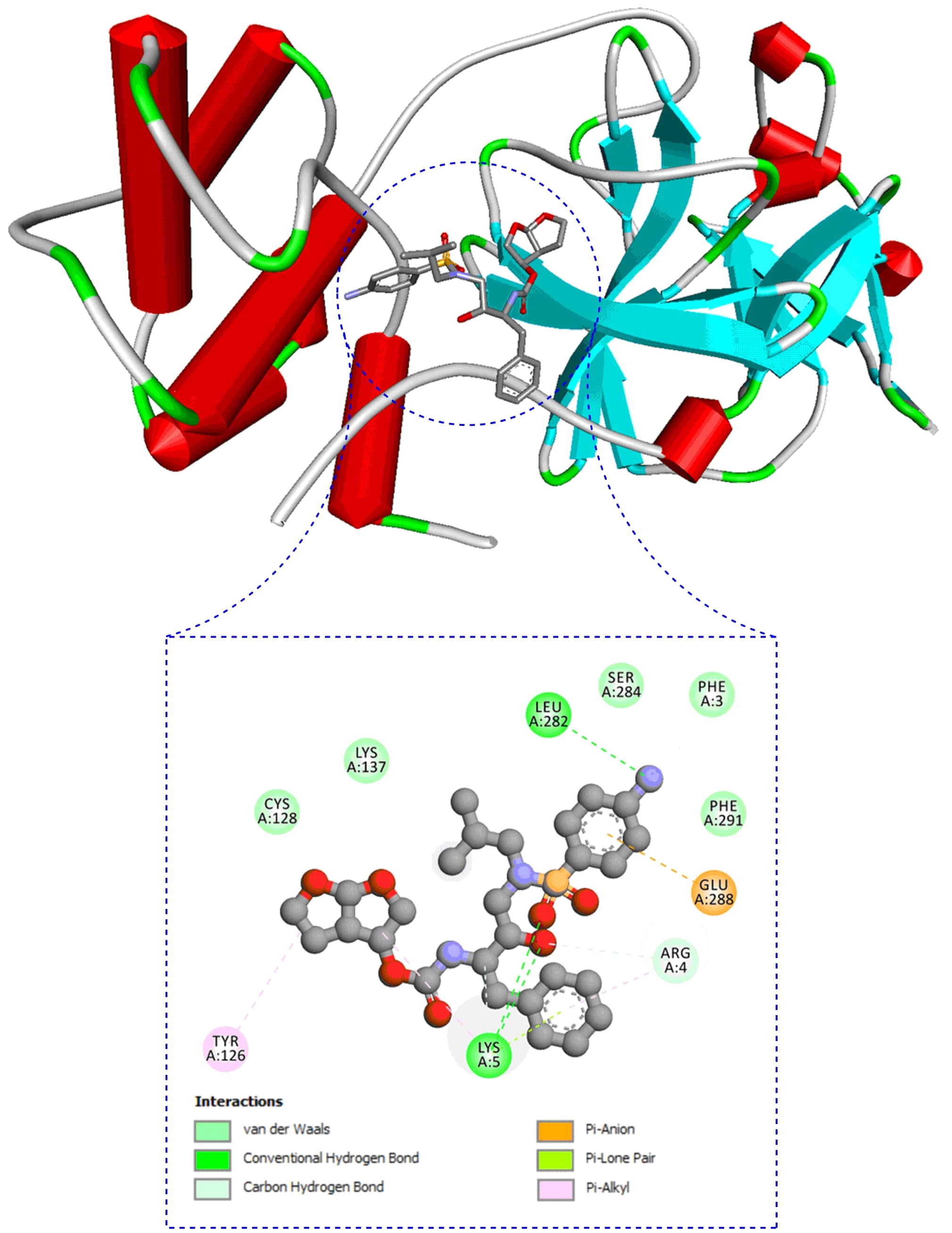 Fig. 3.
Fig. 3.Molecular interaction of second most effective most anti-HIV drug, darunavir with docking score, -10.25 kcal/mol against SARS-CoV-2 protease from docking study.
Likewise, a double docking study between darunavir and VCD-6 and VCD-3 against SARS-CoV-2 main protease (VCD-6 and VCD-3 were re-docked against main protease-darunavir docking complex, individually) was performed. It was discernable that the docking score between darunavir and VCD-6 (ligand-ligand) was only -2.05 kcal/mol, but the score was elevated to -10.76 kcal/mol when dual ligands (protein – ligand + ligand) were used in docking studies. Similarly, the docking score between darunavir and VCD-3 alone was -2.45 kcal/mol and the score was found to be enhanced to -10.39 kcal/mol when dual/second ligands were used in docking studies. As per the present hypothesis, a total docking score of both ligand-ligand and dual docking score of -12.81 and -12.84 kcal/mol were anticipated (Fig. 4). Thus, both VCD-6 and VCD-3 are proposed to be used with darunavir in the combinatorial approach against SARS-CoV-2. According to the structural activity relationship (SAR), VCD-6 was comparatively more active as more hydroxy (-OH) groups support establishing strong-H-bond interaction and sulfate groups support enhancing the biological activity.
 Fig. 4.
Fig. 4.Combinatorial approach of anti-HIV drug with VCD. (A) Darunavir (in red color) with the VCD-6 (in green color). (B) Darunavir (in red color) with the VCD-3 (in green color) at a time against SARS-CoV-2 protease.
Several strategies have been used to screen/filter the potential lead drug
candidates passing through a specific standard parameter at the early stage of drug
discovery. Herein, after docking interaction, we took several ideal drug candidate
selection parameters into account for more evidence of drug-ability at the
primary stage based on their chemical composition. During the selection of most
combinatorial candidates from vitamin C derivatives, all possible biological
activities such as anti-oxidant, anti-inflammatory, immune-stimulant,
vaso-protective, anti-viral and respiratory analeptic properties were carefully
considered and analyzed by the PASS tool (Table 3). From recorded anti-viral
values, only VCD-6 and VCD-2 PASS scores, 0.715
| VCDs | Vaso-protective | Respiratory analeptic | Immuno-stimulant | Anti-oxidant | Anti-inflammatory | Anti-viral |
| VCD-1 | 0.948 |
0.339 |
0.557 |
0.928 |
0.779 |
0.567 |
| VCD-2 | 0.979 |
0.980 |
0.873 |
0.929 |
0.742 |
0.715 |
| VCD-3 | 0.934 |
0.916 |
0.719 |
0.797 |
0.804 |
0.599 |
| VCD-4 | 0.949 |
0.879 |
0.628 |
0.875 |
0.595 |
0.691 |
| VCD-5 | 0.944 |
0.951 |
0.560 |
0.897 |
0.603 |
0.504 |
| VCD-6 | 0.917 |
0.523 |
0.583 |
0.821 |
0.703 |
0.715 |
| VCD-7 | 0.880 |
0.890 |
0.602 |
0.918 |
0.603 |
0.552 |
| VCD-8 | 0.878 |
0.876 |
0.713 |
0.868 |
0.931 |
0.573 |
| VCD-9 | 0.919 |
0.914 |
0.546 |
0.911 |
0.615 |
0.547 |
| VCD-10 | 0.905 |
0.964 |
0.710 |
0.915 |
0.860 |
0.575 |
Furthermore, based on physicochemical and biological properties, the overall
drug-likens score or drug-ability scores of VCDs were recorded (Fig. 5; Table 1).
All VCDs were found to possess a favorable positive rating of a drug-ability
score to be used as a druggable agent. Notably, VCD-1 and VCD-4 showed
comparatively higher drug-ability scores, 0.74 and 0.80, respectively, were
recorded. The drug-ability score of VCD-6 and VCD-3 was 0.24 and 0.34,
respectively. On the other hand, the drug-likeness score of the most effective
anti-HIV drugs, darunavir and tipranavir were recorded to be 0.60 and 0.72,
respectively, as per the tool MolSoft (Table 2). The drug-likeness score depends
on the chemical composition and characteristics within an acceptable range, such
as molecular weight, XlogP value, the number of rotatable bonds, molar
refractivity, and the number of hydrogen bond-acceptors and hydrogen bond-donors,
topological surface area, etc. As per the principle of MolSoft, the drug-likeness scores range within a 0 to 2 considered in the acceptable drug-likeness score, and less than
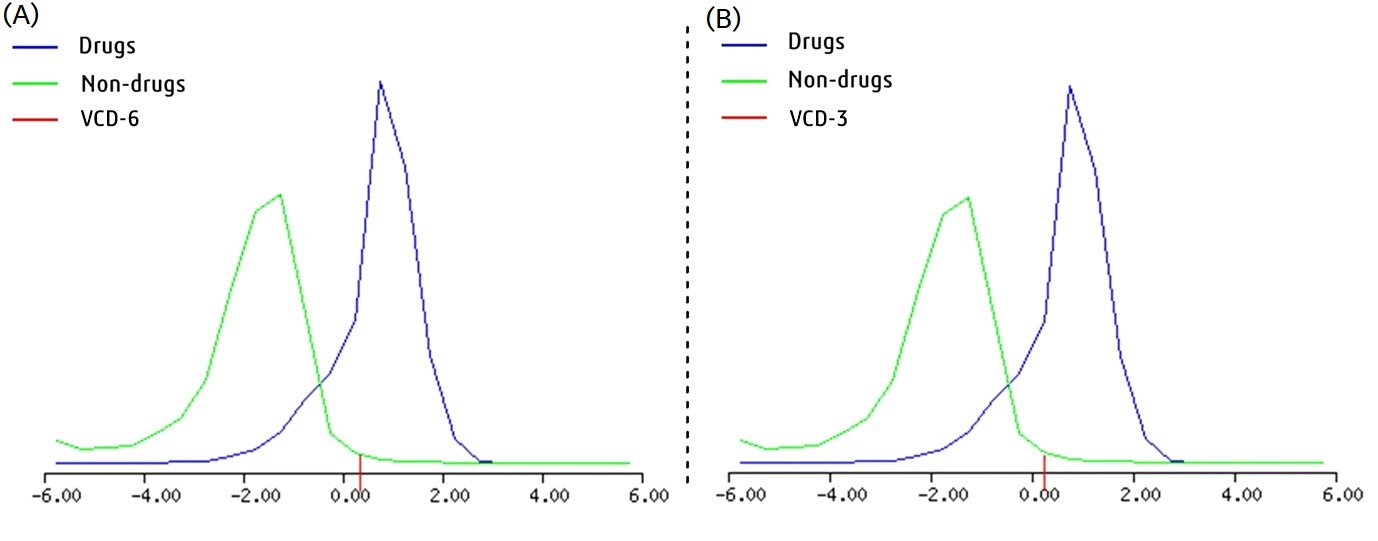 Fig. 5.
Fig. 5.Overall drug likeness score of two effective most VCDs. (A) VCD-6 (0.24). (B) VCD-3 (0.34), respectively.
The pharmacokinetics and toxicity profile also play a crucial role in the late
stage of drug development modules or clinical validation stage before the human
recommendation. Thus, the computational tools can predict the possible
pharmacokinetic and toxicity profiles of any proposed drug molecule. Accordingly,
the used ligands toxicity and the pharmacokinetic profiles were recorded (Tables 4,5).
Results indicate that the VCD-anti-HIV drug combination formulation will make
it safer and balance the toxicity during administration as VCDs showed
potential antioxidative and anti-inflammatory activity. The higher toxicity
class-VI for VCD-6 and VCD-3 also indicated that both are non-toxic and safer
among all VCDs (Table 4). Additionally, the predicted LD
| VCDs | Hepatotoxicity | Carcinogenicity | Immuno-toxicity | Mutagenicity | Cyto-toxicity | Toxicity class |
| VCD-1 | Safest | Safest | Safest | Safest | Moderately safe | V |
| VCD-2 | Safest | Safest | Safest | Safest | Moderately safe | VI |
| VCD-3 | Safest | Safest | Safest | Safest | Moderately safe | VI |
| VCD-4 | Safest | Safest | Safest | Safest | Moderately safe | V |
| VCD-5 | Safest | Safest | Safest | Moderately safe | Moderately safe | V |
| VCD-6 | Safest | Safest | Safest | Moderately safe | Safest | VI |
| VCD-7 | Safest | Moderately safe | Safest | Safest | Moderately safe | V |
| VCD-8 | Safest | Safest | Safest | Moderately safe | Moderately safe | V |
| VCD-9 | Safest | Safest | Safest | Safest | Moderately safe | V |
| VCD-10 | Safest | Safest | Safest | Moderately safe | Moderately safe | V |
| The drugs are classified based on the levels of toxicity as Safest and moderately safe. | ||||||
| Anti-HIV drugs | Hepatotoxicity | Carcinogenicity | Immunotoxicity | Mutagenicity | Cytotoxicity | Toxicity class |
| Amprenavir | Safest | Moderately safe | Safest | Safest | Moderately safe | III |
| ASC09 | Moderately safe | Moderately safe | Toxic/Hazardous | Safest | Moderately safe | III |
| Atazanavir | Moderately safe | Moderately safe | Safest | Moderately safe | Moderately safe | III |
| Darunavir | Toxic/Hazardous | Moderately safe | Moderately safe | Moderately safe | Moderately safe | III |
| Lopinavir | Safest | Safest | Safest | Safest | Safest | V |
| Indinavir | Toxic/Hazardous | Moderately safe | Toxic/Hazardous | Safest | Moderately safe | V |
| Nelfinavir | Moderately safe | Moderately safe | Toxic/Hazardous | Safest | Moderately safe | IV |
| Ritonavir | Toxic/Hazardous | Moderately safe | Safest | Safest | Moderately safe | IV |
| Saquinavir | Moderately safe | Moderately safe | Safest | Safest | Safest | IV |
| Tipranavir | Toxic/Hazardous | Moderately safe | Moderately safe | Safest | Moderately safe | IV |
| The drugs are classified based on the levels of toxicity as Safest, moderately safe and toxic/hazardous. | ||||||
| VCDs | GI-abs. | BBB permit | P-gp substrate | CYP1A2 inhibitor | CYP2C19 inhibitor | CYP2C9 inhibitor | CYP2D6 inhibitor | CYP3A4 inhibitor | Log K |
| VCD-1 | High | No | No | No | No | No | No | No | −8.54 |
| VCD-2 | Low | No | Yes | No | No | No | No | No | −10.55 |
| VCD-3 | High | No | Yes | No | No | No | No | No | −7.48 |
| VCD-4 | Low | No | No | No | No | No | No | No | −9.69 |
| VCD-5 | High | No | No | No | No | No | No | No | −8.61 |
| VCD-6 | Low | No | No | No | No | No | No | No | −9.45 |
| VCD-7 | High | No | No | No | No | No | No | No | −7.70 |
| VCD-8 | High | No | No | No | No | No | No | No | −8.63 |
| VCD-9 | High | No | No | No | No | No | No | No | −8.78 |
| VCD-10 | High | No | No | No | No | No | No | No | −8.39 |
| BBB, blood-brain barrier; GI-abs., gastrointestinal absorption Log K | |||||||||
 Fig. 6.
Fig. 6.Overall predicted pharmacokinetics reports of two effective most VCDs. (A) VCD-6. (B) VCD-3.
The structural stability of selected two docking complexes was analyzed through the root mean square deviation (RMSD)-protein backbone, root mean square fluctuation (RMSF)-C-alpha and Radius of gyration (Rg) of protein by MD simulation in 50 ns time scale (Figs. 7,8). The elucidated RMSD plot for both ‘protease-darunavir’ and ‘protease-VSD-6’ complexes showed a continuous deviation in the backbone protein during 50 ns. Mainly, darunavir holds some stability in stating within 20 ns, while both are in the same fluctuation place at the end, after 45–50 ns (Fig. 7A). At the same from each ligand flexibility study, VCD-6 maintained more stability than darunavir throughout 50 ns (Fig. 7B). Overall, VCD-6 showed comparatively more stability than darunavir from RMSD analysis. Rg-plots and RMSF-plots also showed that both complexes were similar fluctuation with 50 ns MD simulation. Additionally, both ligands could therapeutically potential towards block the viral function through solid H-bond interactions with SARS-CoV-main protease (PDB ID: 6Y84) from the MD simulation (Fig. 8).
 Fig. 7.
Fig. 7.Conformational stability in the form of RMSD-score. (A) Stability of protease-darunavir’ and protease-VCD-6’ docking complexes. (B) Individual ligand stability in the docked complexes within 0–50 ns in the overlayed figure.
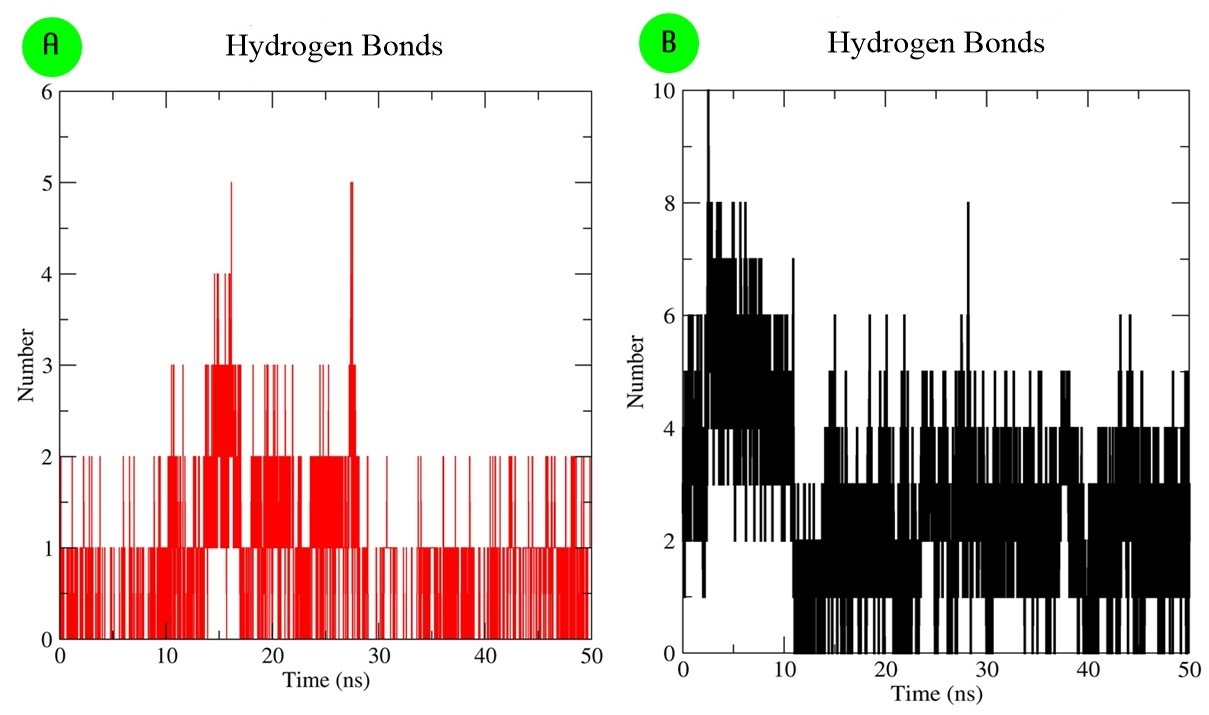 Fig. 8.
Fig. 8.Interaction stability analyses in the form of H-bond. (A) H-bond interaction of ‘protease-darunavir’ docking complex. (B) H-bond interaction of ‘protease-VCD-6’ docking complex during 50 ns.
The advanced computation program presently plays an ideal platform to accelerate the drug discovery process with minimal resources and time during the selection of lead drug candidates [26, 28, 29]. Indeed, advanced computational intelligence is able to mimic the entire human living system through coding/programming languages. Molecular docking is one of the efficient computational platforms used for locating active therapeutic agents, based on the interaction between the targeted enzyme and ligand complexes [25, 28]. Molecular docking has also been applied since the last decade to efficiently screen out several potential drug molecules than traditional wet-laboratory approaches. Theoretically, docking score is the sum of the individual molecular interaction of the ligand with the target protein, where types of bonds (conventional H-bond is stronger than other bond formation), bond-length (lesser in the distance (Å) consider as more in strength) [25, 26, 27]. For example, VCD-6 showed an effective docking score as most of the interactions in their complex formed with H-bonds with minimum bond-length (Å) (Table 1). Thus, it will continue to play an essential role in early drug discovery before experimentation [28, 29, 30]. Several pharmaceutical industries and exclusive drug research laboratories/stations are also using this advanced technique in novel drug development cascades [29, 31].
On the other hand, natural products are always vital for nutraceuticals, pharmaceutical supplements, especially against infectious diseases [32, 33]. Identified several unique secondary metabolites from different medicinal and aromatic plants are continuously used in different ways for control of several human health ailments [20, 32, 33]. Indeed, natural products have been in use in several developed western countries as Complementary and Alternative Medicine (CAM) [20, 34, 35]. Approximately 48% of the population in Australia, 70% in Canada, 42% in the USA, 38% in Belgium and 75% in France communities use phytoextract as CAM in day-to-day life [36]. Indeed, WHO also emphasizes developing or locating natural-product(s) based drug candidates for mainstream use. Simultaneously, ascorbic acid or vitamin C derivatives, having a wide range of biological activity against respiratory tract infection, with anti-bacterial, anti-viral and potent immune-stimulant activities [37, 38, 39, 40]. Recently, some non-clinical reports also indicate that vitamin C has potent anti-SARS-CoV-2 activity and is now under clinical validation too (trail no. NCT04323514). On the other hand, anti-HIV drugs are the most suitable therapeutic choice against SARS-CoV-2, as previously exhibited positive response against SARS and MARS on a repurposing basis with severe side effects from long-term practice. Thus, the present attempt with a suitable anti-oxidative and immune-stimulant natural regimen based-combinatorial approach towards control of SARS-CoV-2 may minimize the host toxicity.
In the present scenario, finding a newer drug candidate is a challenge. Most pharmaceutical companies face the same problem as selected lead candidates disappointment in clinical trials due to lack of drug-likeness property, even reported active in preliminary in vitro and in vivo testing [33, 36, 41]. Thus, biological activity is a primary parameter for to establishing a lead candidate and several other standard parameters should be followed during clinical trial investigation for recognition in mainstream medicine. The advanced artificial inelegance platform analyzed this parameter for any desired or unknown chemical through a data mining procedure. Hence, the drug-ability investigation based on high throughput computational screening is more beneficial to keep a suitable drug lead candidate at the early stage [23, 24, 25, 26, 27, 28].
Several mainstream medicines come from natural resources or contain active biological scaffolds/constituents and require continuous exploration of more natural products to utilize in mainstream medicine to fulfill the unavailability of drugs in an emergency [32, 42]. The current bioassay-guided plant extraction and characterization methods have isolated many active secondary metabolites for therapeutic uses. At the same time, advanced medicinal chemistry and bioinformatics tools currently help locate active candidates targeting any specific pathogen/disease to accelerate and give enough reports for early drug discovery [41, 42]. Under the emergency, this selected combination could be the right option than any non-clinical hit-and-trial prescription to treat SARS-CoV-2 infected patients. Notably, using two anti-HIV drugs might be associated with a higher risk of hepatic and liver injury to patients. Thus, potent vitamin C could be the right candidate with an anti-HIV drug to manage side effects for its anti-oxidative, anti-inflammatory potency, immune-stimulant, with anti-viral efficacy.
Today, COVID-19 is the most significant pandemic in the 21st century caused by the aggressive mutant SASR-CoV-2 strains in a different interval that the world has been observing since the end of 2019. The lockdown and social distancing as preventive actions to break/pause the viral cycle within the community. However, potential and exclusively vaccine/therapeutic is essential to control the gruesome SARS-CoV-2 strain [43, 44, 45]. The combinatorial drug therapy as projected herein using existing medicines and the knowledge of traditional Indian medicine/Ayurveda and traditional Chinese medicine is ingenious and motivated attempts towards control of SARS-CoV-2 and reduces adverse effects from repurposing drugs.
From the management and treatment points of view, physicians used several alternative combinations of existing anti-viral, anti-inflammatory and anti-malarial drugs with less clinical experience to tackle the situation of SASR-CoV-2 infection on a case-by-case basis. Still, the post-treatment side effects of two mainstream medicines used in combination drug therapy are under debate. Thus, the present study intends to find a possible potent combinatorial approach with an existing anti-HIV drug, darunavir with a vitamin C derivative against SARS-CoV-2. The potent combination was verified in a cost-effective drug development platform and suggested based on advanced PASS prediction, molecular docking, toxicity-pharmacokinetic profile prediction and MD simulation analyses. It could be a dominant synergistic formula of a natural derivative with an anti-HIV drug to control COVID-19 infection and is expected to produce negligible toxicity due to the presence of anti-oxidative, anti-inflammatory and immune-stimulant natural vitamin C derivative with darunavir. In an emergency, the computer-aided drug design approach utilizing present clinical evidence and genomic information on SARS-CoV-2 gives hints to investigate natural products for its treatment. Finally, verifications of the essential features of prospective drugs loom large ordinarily to a traditional pharmacologist, who would blithely accept the results of bioinformatics analyses for promotion towards drug development screening.
ADMET, absorption, distribution, metabolism, excretion and toxicity; CAM,
complementary and alternative medicine; COVID-19, coronavirus-2019 disease; FDA,
food and the drug administration; H-bond, hydrogen bond; HIV, human
immunodeficiency virus; LD
SSS and AS designed and hypothesized the concept. AS carried out the computational work under the guidance of SSS. SSS and AS drafted the MS. BP and MP carefully reviewed and commented on the final MS. All authors contributed to manuscript revision, read, and approved the submitted version.
Not applicable.
We are thankful to the Dean, Institute of Medical Sciences & SUM Hospital, Siksha ‘O’ Anusandhan Deemed to be University and the Director, ICMR-Regional Medical Research Centre, Bhubaneswar, for encouragement during the preparation of the MS and special thanks to the World Health Organization for updated information on SARS-CoV-2. The entire work is dedicated to science and all heroes (doctor, nurse, pharmacist, media, police, soldier, and associated leader) who fight against SARS-CoV-2 for us.
This work is sincerely acknowledged to S‘O’A University in-house Ph.D. fellowship support to Miss. Alaka Sahoo (Biotechnology Regd. No.: 1981611009) and ICMR-Post Doctoral Fellowship award to Dr. Shasank S. Swain (No.3/1/3/PDF[21]/HRD-2019-2) by the Indian Council of Medical Research, Govt. of India, New Delhi.
The authors declare no conflict of interest.
