†These authors contributed equally.
Academic Editor: Josef Jampilek
Background: Hepatocellular carcinoma (HCC) is a common clinical malignant disease and the second leading cause of cancer-related death worldwide. Dendrobium is a commonly applied nourishing drug in traditional Chinese medicine. Gigantol is a phenolic compound extracted from Dendrobium. The compound has attracted attention for its anticancer effects. However, the mechanism of gigantol in HCC has not been extensively explored. Methods: Potential targets of gigantol were predicted by SwissTargetPrediction. HCC-related genes were obtained from the GeneCards, Online Mendelian Inheritance in Man (OMIM), Pharmacogenetics and Pharmacogenomics Knowledge Base (PharmGKB), Therapeutic Target Database (TTD) and DrugBank databases. The “gigantol-target-disease” network was constructed using Cytoscape software. Protein interaction network analysis was performed using STRING software. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were executed utilizing the R package to explore the possible regulatory mechanisms of gigantol in HCC. To authenticate the role of gigantol in HCC, Cell Counting Kit-8 (CCK-8) assay, 5-ethynyl-2’-deoxyuridine (EdU) assay, wound healing assay, Matrigel invasion assay and Western blot were performed. Results: Three core genes were screened from 32 closely linked genes. Pathway analysis yielded many signaling pathways associated with cancer. The CCK-8 assay and EdU assay indicated that gigantol suppressed the growth of HCC cells. The wound healing assay and Matrigel invasion assay showed the inhibition of migration and metastasis of HCC cells by gigantol. We verified from molecular docking and protein level that gigantol can exert regulatory effects through three targets, ESR1, XIAP and HSP90AA1. Furthermore, Western blot results tentatively revealed that gigantol may inhibit HCC progression through the HSP90/Akt/CDK1 pathway. Conclusions: Our results confirms anti-HCC proliferation activity of gigantol through PI3K pathway described in existing literature by different experimental approaches. Furthermore, it has discovered other proteins regulated by the drug that was not previously reported in the literature.These findings provide potential molecular and cellular evidence that gigantol may be a promising antitumor agent.
Hepatocellular carcinoma is currently the fifth most common malignancy worldwide and the second leading cause of death from cancer [1]. The pathological types of primary liver cancer mainly include hepatocellular carcinoma, intrahepatic cholangiocarcinoma, and mixed hepatocellular carcinoma, of which HCC accounts for approximately 85%–90%. HCC mostly emerges from chronic liver disease or cirrhosis and has an insidious onset. Early symptoms are not always obvious, so most patients are diagnosed at a late stage, resulting in a very poor overall prognosis. The treatment of HCC emphasizes an integrated, multidisciplinary model to improve patient survival. For inoperable HCC, radiation therapy (radiotherapy) [2, 3], radiofrequency ablation (RFA) [4, 5, 6, 7, 8], transarterial chemoembolization (TACE) [9, 10, 11], and systemic therapy are all effective treatment options [12, 13, 14, 15, 16]. Therefore, the study of valuable therapeutic agents will help improve the clinical outcome of patients.
Dendrobium is listed in Shen Nong Ben Cao Jing (The Divine Husbandman’s Classic of Materia Medica) and is classified as a top quality plant as a perennial green herb of the orchid family. The Chinese Pharmacopoeia records that Dendrobium is cold in nature, sweet, light and salty in taste, and enters the stomach, lung and kidney meridians. Due to benefiting the stomach and generating fluid, nourishing Yin and clearing heat. Research has confirmed that many compounds in Dendrobium, especially the bibenzyl and phenolic compounds, have significant antitumor activity and antimultidrug resistance and have inhibitory effects on many human tumor cell lines [17]. Gigantol is a typical phenolic compound in Dendrobium, and some studies have shown that gigantol can inhibit the oxidative stress induced by high sugar and may have a protective effect in the development of diabetic retinopathy, which may help reduce neovascularization [18]. Gigantol inhibits stem cell protein kinase B (Akt) signaling in cancer cells by reducing lung cancer activity, thereby reducing the cellular levels of pluripotency and the self-renewal factors Oct4 and Nanog [19]. Gigantol dramatically reduces the viability of lung cancer cells in the isolated state [20]. Therefore, gigantol may be developed as a therapeutic agent for cancer.
Network pharmacology is an effective method to study the multicomponent and multitarget mechanism of Chinese herbal compounds and is holistic, systematic and connected and coincides with the theory of a holistic view, diagnosis and treatment, and prescription dispensing in Chinese medicine [21, 22, 23, 24]. In this study, we aimed to construct a “compound-target-disease” network based on network pharmacology and explore the molecular mechanisms and pathways of gigantol in the treatment of HCC through molecular docking and in vitro validation.
Structural information on gigantol (CAS:67884-30-4) was obtained from NCBI
PubChem (https://pubchem.ncbi.nlm.nih.gov/) [25]. Swiss absorption, distribution,
metabolism and excretion (SwissADME), was used to identify the bioavailability and
metabolism of gigantol [26]. Additionally, the protein targets of this compound
were retrieved from SwissTargetPrediction with a probability value
The common targets were uploaded to the STRING database (https://string-db.org/), and the protein interaction network was constructed with the following conditions: the species was “Homo sapiens”, the minimum interaction score (medium confidence) was 0.4, and the free protein sites were removed [32]. The obtained protein interaction networks were imported into Cytoscape 3.8.2 software (Institute of Systems Biology, Boston, MA, USA) for visualization and processing analysis using the Network Analyzer module [33]. Finally, the core genes were screened by the cytoNCA plug-in. The cytoNCA plug-in performs topology analysis with a 2-fold average degree value as a filtering condition.
GO and KEGG pathway enrichment analysis were performed using the R
package (“clusterProfiler” “org.Hs.eg.db”
“enrichplot”
“ggplot2” “pathview”).
The significance values for GO terms and KEGG pathways were set at FDR
The two-dimensional structure of gigantol were available from the PubChem website in structured data format (SDF) format [25]. The 3D structure of the core target protein was downloaded from the PDB database (https://www1.rcsb.org/) and saved in PDB format [34]. Water molecules and small molecule ligands were removed using PyMOL software (Schrödinger, New York, USA) and hydrogenated using AutoDockTools software (Scripps Research, La Jolla, CA, USA) (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC) [35]. Molecular docking of the receptor and ligand was performed using Vina 1.1.2 (Scripps Research, La Jolla, CA, USA) [36]. The binding activity was evaluated by docking score values.
The human hepatocellular carcinoma cell lines Hep3B, SMMC-7721, and HCC-LM3 were
obtained from the Chinese Academy of Sciences (Shanghai, China). Cells were
cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco, Grand Island, NY, USA)
supplemented with 10% FBS (Gibco, Grand
Island, NY, USA) and 100 U/mL Penicillin/Streptomycin (Gibco, Grand Island, NY,
USA) in a 5% CO
A CCK-8 assay kit was used to evaluate cell viability following the
manufacturer’s instructions (HY-K0301,
MedChemExpress, Monmouth Junction, NJ, USA).
Cells in the logarithmic growth phase were inoculated into 96-well plates (7
A EdU detection kit (Us Everbright, Suzhou, Jiangsu, China) was used, as stated
by the manufacturer’s instructions, to identify the proliferating cells. SMMC-7721
and Hep3B were intervened with different concentrations of gigantol for 24 h
after apposition. Appropriate amounts of cells were inoculated in 96-well plates
and cultured in the logarithmic growth phase. EdU solution was diluted with
complete medium at a 1000:1 ratio, 100
Hep3B and SMMC-7721 cells were seeded onto cell inserts (Culture-Insert 4 well,
ibidi, Gräfelfing, Germany) in a 12-well plate at a density of 5
For the transwell invasion assay, matrigel chambers (BD Biosciences, San Jose,
CA, USA) were carried out according to manufacturer’s instructions. Matrigel was
melted at 4
Cells in logarithmic growth phase were seeded in 6-well plates at No. 1
GraphPad Prism 8.0 software (GraphPad
Software, San Diego, CA, USA) for Windows was used for statistical analysis.
Statistically significant differences were calculated using ANOVA. A
p-value
The SwissADME results indicated that gigantol not only undergoes passive gastrointestinal absorption but also crosses the BBB (Table 1) (http://www.swissadme.ch/index.php). A total of 102 gigantol-associated targets were retrieved (Supplementary Table 1). Subsequently, a total of 1184 hepatocellular carcinoma related target genes were compiled from the OMIM, PharmGKB, GeneCards, TTD and DrugBank databases (Fig. 1A). Thus, we integrated the drug targets and disease targets using the R package “Venn”. After eliminating duplicates, we identified 32 common targets, which are shown in Fig. 1B.
| Property | Parameter |
| MW | 274.31 g/mol |
| TPSA | 58.9 Ų |
| XLogP3-AA | 2.7 |
| H-bond Donor Count | 2 |
| H-bond Acceptor Count | 4 |
| Rotatable Bond Count | 5 |
| GI absorption | High |
| BBB permeant | Yes |
| MW, molecular weight; TPSA, Topological polar surface area; XLogP3-AA, computed oc-tanol/water partition coefficient; BBB permeant, blood brain barrier permeant. | |
 Fig. 1.
Fig. 1.Analysis of the predicted genes. (A) Intersection of HCC target genes predicted by five types of databases. (B) Venn diagram of HCC-related genes and gigantol target genes. Using R packages (“Venn”).
The 32 intersecting targets were imported into the STRING database to obtain the protein interaction network and the corresponding network (Fig. 2A–B). The data were loaded into Cytoscape for visual topological analysis, and 32 nodes with 148 edges and an average degree value of 9 were obtained, in which ESR1, ERBB2, HSP90AA1, MAPK8, CDK4, XIAP, RPS6KB1, PTGS2, CDK1, IGF1R, ABL1, PARP1, and CHEK1 were selected (Fig. 2C) (Table 2). Then, we further analyzed the high score proteins using the cytoNCA plug-in and obtained 13 nodes with 58 edges and an average degree value of 8.9. Three targets, HSP90AA1, XIAP, and ESR1, were identified as occupying a central position in the PPI network and were expected to serve as key core targets (Fig. 2D).
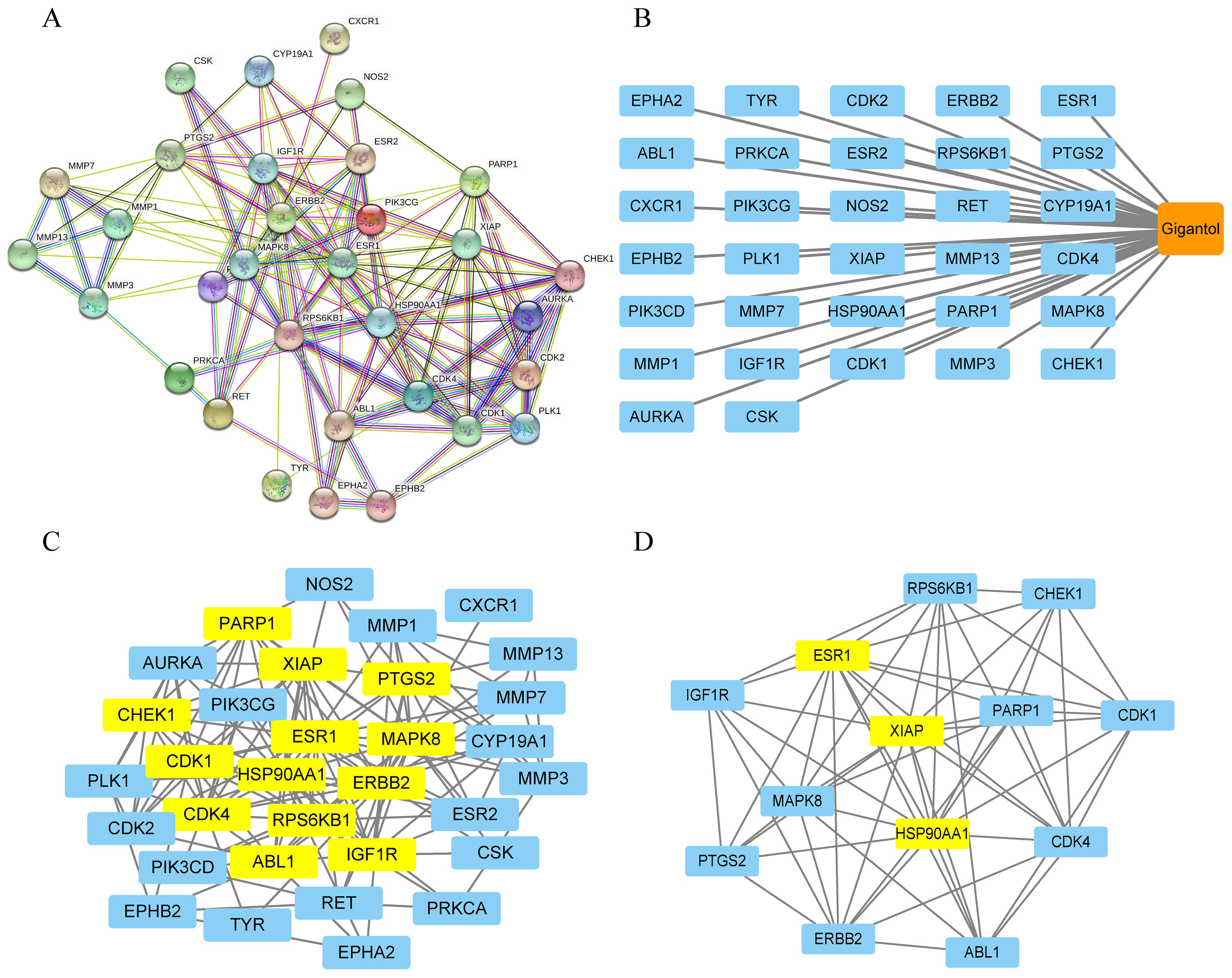 Fig. 2.
Fig. 2.Network pharmacology of gigantoland HCC. (A) The PPI network of common genes. (B) The “HCC-targets-gigantol” network. (C) Visualization of the first 13 core genes was obtained using R software. (D) The top 3 potential effective targets in core genes. Obtained by Cytoscape and cytoNCA plug-in analysis for mapping.
| Name | Betweenness | Closeness | Degree | Eigenvector | LAC | Network |
| ESR1 | 119.458775 | 0.756097561 | 21 | 0.334978998 | 8.571428571 | 17.59735988 |
| ERBB2 | 193.2986109 | 0.738095238 | 20 | 0.266285211 | 5.7 | 13.74281303 |
| HSP90AA1 | 91.99843591 | 0.720930233 | 19 | 0.311388731 | 8.210526316 | 15.2760101 |
| MAPK8 | 86.93683 | 0.688888889 | 17 | 0.249040633 | 6.588235294 | 13.0373002 |
| CDK4 | 47.88048556 | 0.645833333 | 15 | 0.253567874 | 7.466666667 | 11.83308358 |
| XIAP | 18.81066257 | 0.645833333 | 14 | 0.271532387 | 8.571428571 | 10.71085859 |
| RPS6KB1 | 35.45680286 | 0.62 | 13 | 0.220462099 | 5.846153846 | 7.91991342 |
| PTGS2 | 35.8962606 | 0.62 | 13 | 0.197408468 | 6.307692308 | 10.29350649 |
| CDK1 | 15.60249175 | 0.58490566 | 12 | 0.217495233 | 7.833333333 | 9.890909091 |
| IGF1R | 26.57395043 | 0.607843137 | 12 | 0.197221518 | 6 | 8.968181818 |
| ABL1 | 24.98813008 | 0.607843137 | 11 | 0.204987556 | 6 | 7.655555556 |
| PARP1 | 15.52287496 | 0.607843137 | 11 | 0.214561418 | 6.909090909 | 8.319444444 |
| CHEK1 | 8.966233354 | 0.574074074 | 11 | 0.204811394 | 7.454545455 | 8.946031746 |
| CDK2 | 1.773809524 | 0.553571429 | 10 | 0.195786715 | 7.8 | 8.968253968 |
| AURKA | 0.571428571 | 0.543859649 | 9 | 0.179965124 | 7.555555556 | 8.607142857 |
| PLK1 | 7.201731602 | 0.534482759 | 8 | 0.147147462 | 5.5 | 6.392857143 |
| ESR2 | 2.252380952 | 0.563636364 | 8 | 0.158232242 | 5.75 | 6.8 |
| MMP7 | 5.584594817 | 0.553571429 | 7 | 0.109712064 | 5.142857143 | 6.333333333 |
| MMP1 | 5.584594817 | 0.553571429 | 7 | 0.109712064 | 5.142857143 | 6.333333333 |
| MMP3 | 10.06937866 | 0.508196721 | 7 | 0.093782939 | 4 | 5 |
| PIK3CG | 4.400582338 | 0.516666667 | 6 | 0.10157793 | 3 | 3.75 |
| CYP19A1 | 1.203361345 | 0.534482759 | 6 | 0.117587544 | 4 | 4.8 |
| RET | 21.12384817 | 0.553571429 | 6 | 0.087166645 | 1.333333333 | 1.65 |
| EPHB2 | 5.642857143 | 0.442857143 | 5 | 0.060849536 | 1.2 | 1.583333333 |
| PIK3CD | 2.692929293 | 0.492063492 | 5 | 0.072902597 | 2.4 | 3 |
| MMP13 | 0 | 0.449275362 | 5 | 0.063357756 | 4 | 5 |
| EPHA2 | 4.434199134 | 0.516666667 | 4 | 0.070508458 | 2 | 2.666666667 |
| NOS2 | 0.333333333 | 0.492063492 | 4 | 0.081238903 | 2.5 | 3.333333333 |
| PRKCA | 3.74042624 | 0.5 | 4 | 0.059561979 | 0.5 | 0.666666667 |
| CSK | 0 | 0.46969697 | 3 | 0.059485048 | 2 | 3 |
| TYR | 0 | 0.476923077 | 2 | 0.043449122 | 1 | 2 |
| CXCR1 | 0 | 0.430555556 | 1 | 0.022233263 | 0 | 0 |
GO enrichment analysis was performed on 32 intersecting targets using the R language package. A total of 707 biological process (BP) entries were obtained, involving peptidyl-serine modification, regulation of cell cycle G2/M phase transition, peptidyl-serine phosphorylation, regulation of protein kinase B signaling, signal transduction in response to DNA damage and other processes. Thirty seven cellular component (CC) entries were obtained, involving the pigment granule membrane, nuclear chromosome, telomeric region, cyclin-dependent protein kinase holoenzyme complex, and other components. Forty two molecular function (MF) entries involved nuclear receptor binding, transmembrane receptor protein tyrosine kinase activity, and cyclin-dependent protein kinase activity. The top 10 entries of the 3 cluster analyses were plotted according to the number of genes sorted (see Fig. 3A). KEGG enrichment analysis was performed on 32 intersecting targets with the R package. A total of 106 entries were obtained, and the results suggested that genes were predominantly enriched in the PI3K-Akt signaling pathway, endocrine resistance, ErbB signaling pathway, IL-17 signaling pathway, HIF-1 signaling pathway, and hepatocellular carcinoma, as shown in Fig. 3B (Supplementary Table 2). The results clearly revealed that the PI3K-Akt signaling pathway was more abundant among HCC-related pathways and had a critical role in the treatment of HCC (Fig. 4).
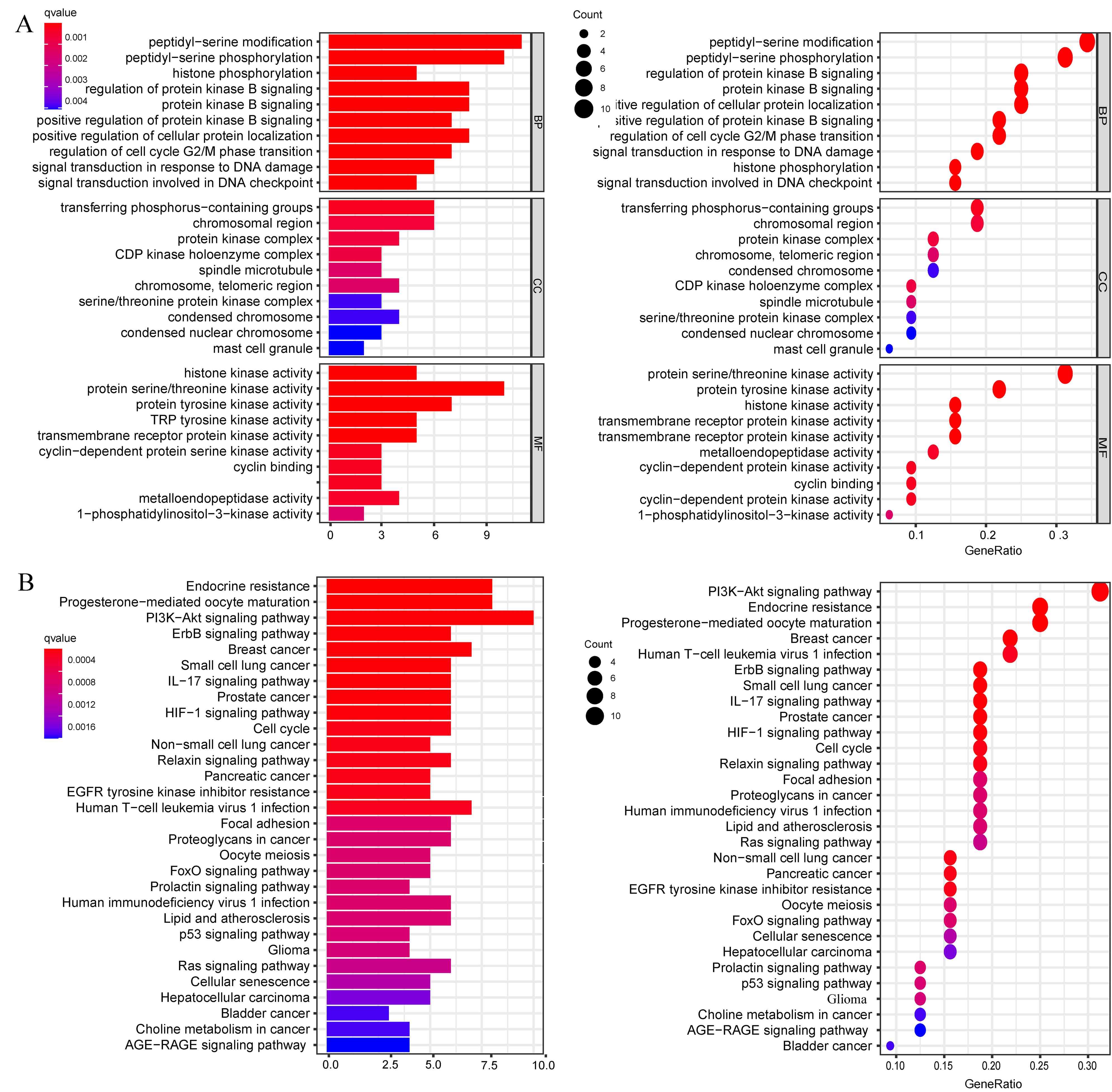 Fig. 3.
Fig. 3.Enrichment analysis of the 32 genes by GO and KEGG. (A) GO functional annotation of 32 genes of gigantol. BP, CC and MF were analyzed using R software. (B) KEGG pathway analysis of 32 genes and column plot for top 30 pathways. GO and KEGG analysis were performed using the R package (GO, KEGG: “clusterProfiler”, “org.Hs.eg.db”, “enrichplot”, “ggplot2”).
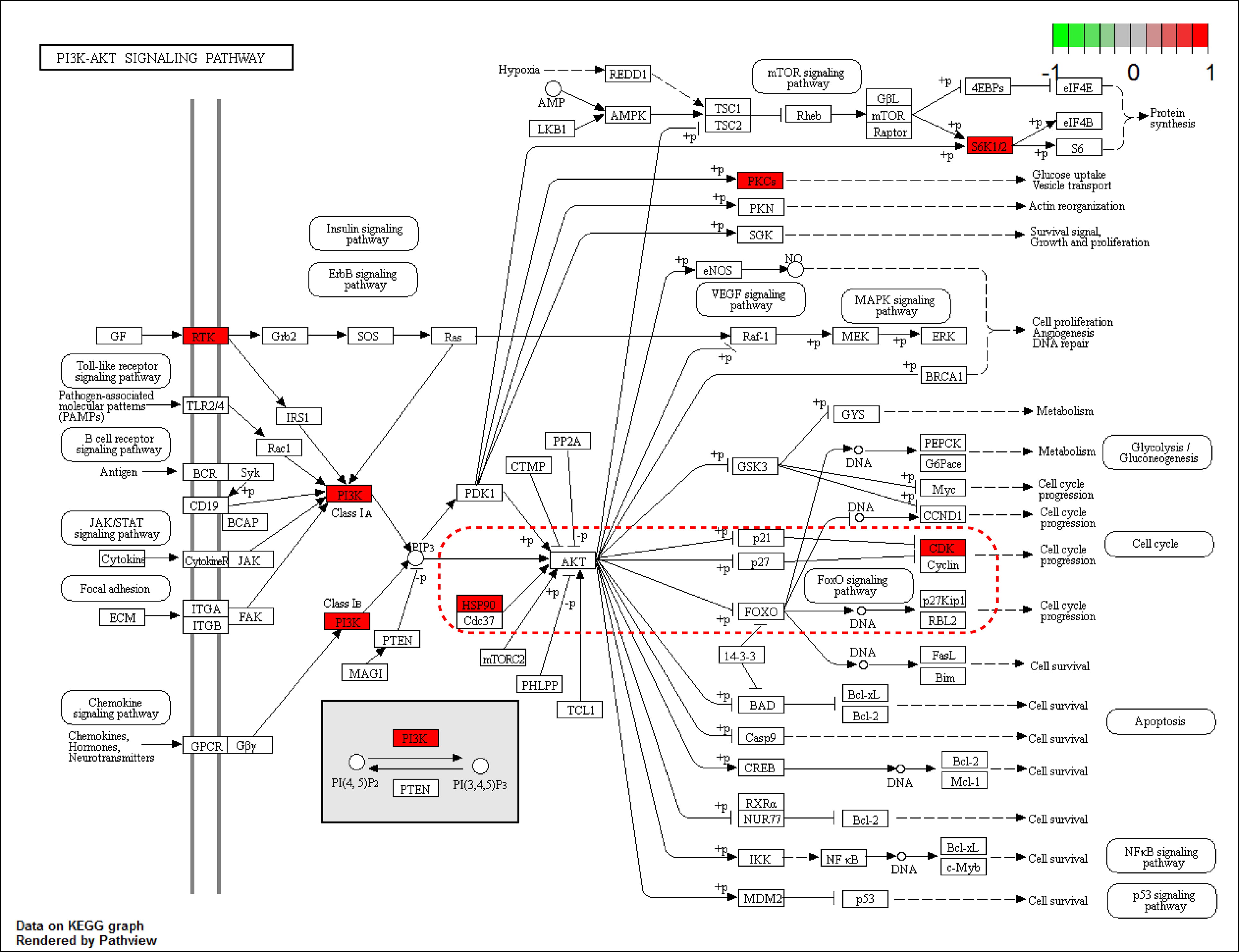 Fig. 4.
Fig. 4.The PI3K-Akt signaling pathway. The nodes marked in red are target genes. The red dashed box highlights our predicted key drug targets in the PI3K-Akt pathway. HSP90 may be one of the targets for the action of gigantol. HSP90/Akt/CDK1 is the likely downstream pathway of interest. Using R packages (“pathview”).
Whether a small molecule can bind to a large protein is mainly evaluated by the
binding energy, which if it is less than 0, the ligand and the receptor can bind
spontaneously. Smaller values indicate stronger binding capacity where the active
ingredient binds to the receptor more easily. In this experiment, the average
docking score of gigantol was used as the threshold value. The binding energy of
the target protein to small molecules with a threshold value less than –5
kcal/mol was calculated. Three proteins were successfully docked with gigantol.
The molecular docking results revealed that the binding energies of gigantol to
the proteins ESR1, HSP90AA1, and XIAP were –6.5, –6.3, and –6.2 kcal/mol,
respectively, indicating that the three molecules were highly active (Table 3).
Affinity values
| Targets | PDB ID | Grid center | Npts | Spacing | Binding energie | ||||
| ESR1 | 2JFA | –35.047 | 17.223 | –4.757 | 40 | 40 | 40 | 1.0 | –6.5 |
| HSP90AA1 | 3T0H | –0.802 | –14.335 | –20.396 | 40 | 40 | 40 | 1.0 | –6.3 |
| XIAP | 4MTZ | 9.296 | –0.371 | 17.785 | 40 | 40 | 40 | 1.0 | –6.2 |
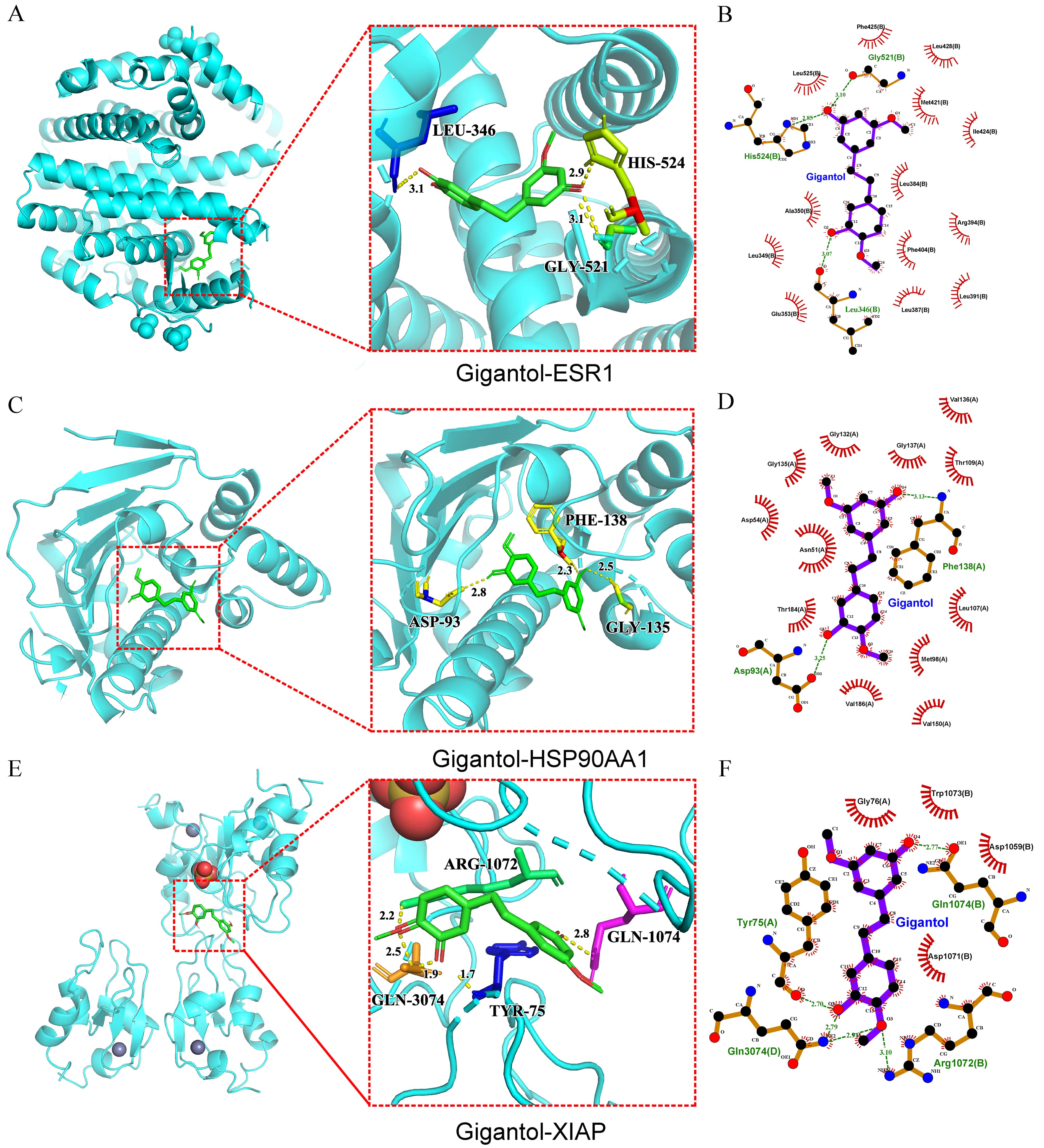 Fig. 5.
Fig. 5.Molecular docking. (A) 3D structural diagrams of the interactions between ESR1, HSP90AA1 (C), XIAP (E) with gigantol, respectively. (B) Plot of the distribution of the interaction forces between ESR1, HSP90AA1 (D), XIAP (F) and gigantol, individually.
The inhibition rates of gigantol on Hep3B, SMMC-7721, and HCC-LM3 cells are
shown in Fig. 6A. For SMMC-7721 cells, the IC
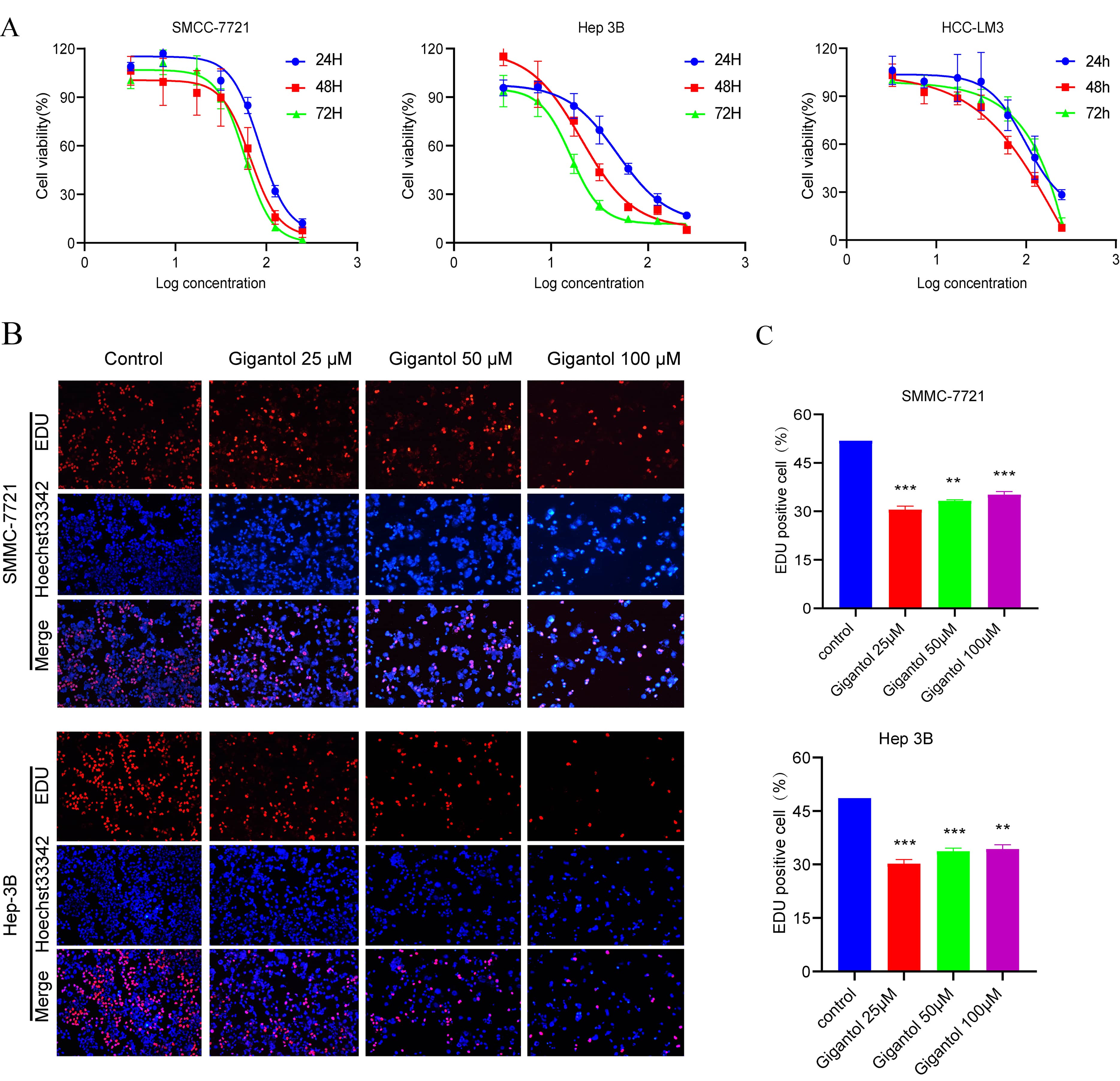 Fig. 6.
Fig. 6.Gigantol inhibits the proliferation of HCC cells. (A) The
viability of SMMC-7721, Hep3B and HCC-LM3 cells treated with different
concentrations of gigantol for 24 h, 48 h and 72 h was examined by CCK-8 assay.
(B) The multiplicative capacity of SMMC-7721 and Hep 3B cells was decreased after
different concentrations of gigantol intervention for 24 h. (C) Bar graphs show the
percentage of EdU-positive cells in SMMC-7721 (p
As shown in Fig. 7A–B, wound healing assays demonstrated that the scratches of gigantol treated Hep3B and SMMC-7721 cells were significantly larger than those of the control cells after 24 h. Additionally, the Matrigel invasion assay demonstrated that gigantol inhibited the invasion rate of Hep3B and SMMC-7721 cells compared to the control group (Fig. 7C–D). The aforementioned results demonstrated that gigantol could effectively inhibit the migration and invasion of HCC cells.
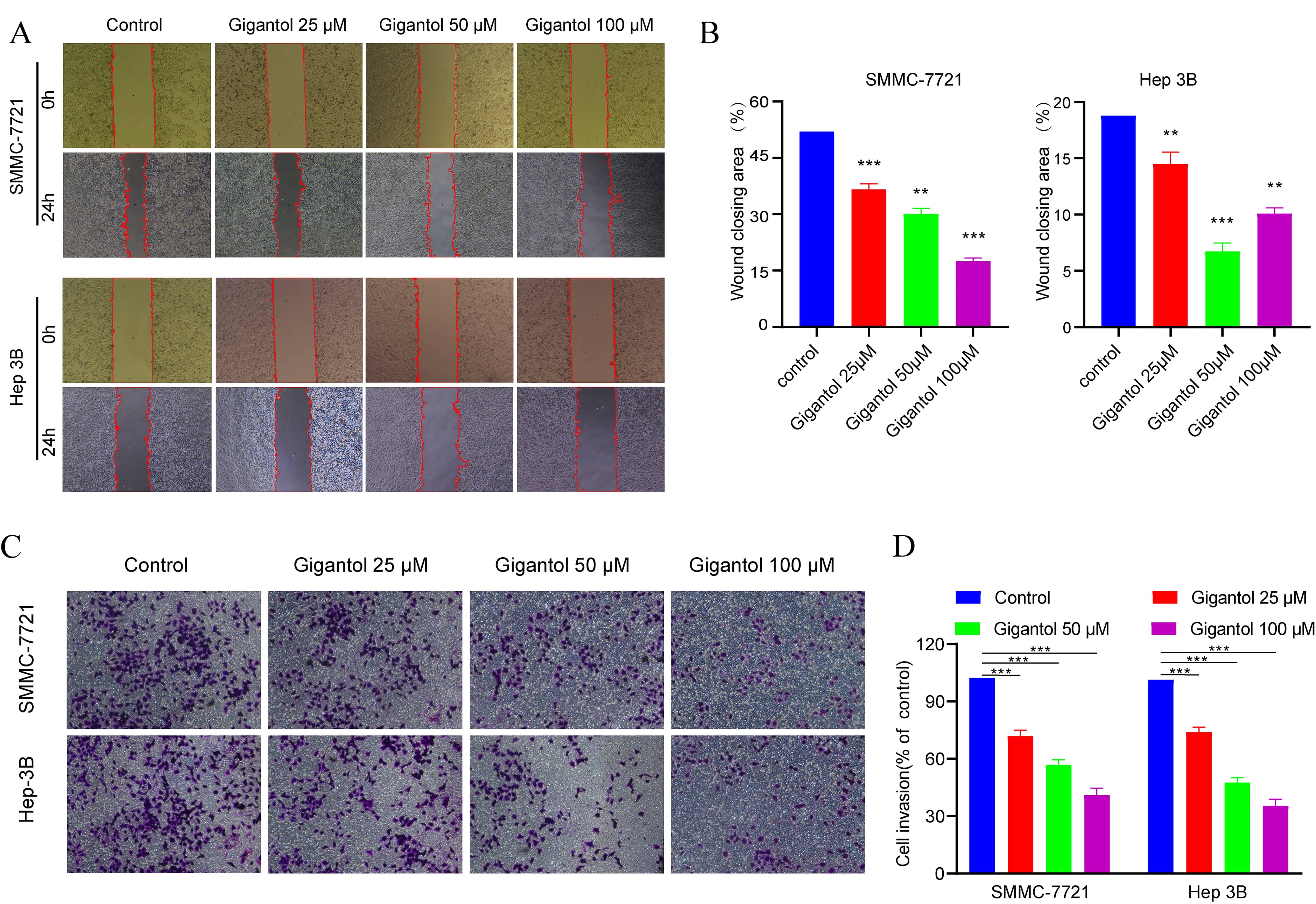 Fig. 7.
Fig. 7.Gigantol suppressed migration and invasion of cells. (A) The
representative micrographs of “wound healing assay” of SMMC-7721 cells and Hep3B
cells after 24 h are shown. (B) Bar graphs show the percentage of the area for
the scratched region in SMMC-7721 (p
To further determine the efficacy of gigantol, we assayed proliferation,
migration and invasion related biomarkers. Following gigantol intervention, PCNA
and MCM2 secretion was significantly diminished in the experimental group
compared to the control group. Stimulation with different concentrations of
gigantol (0, 25, 50, 100
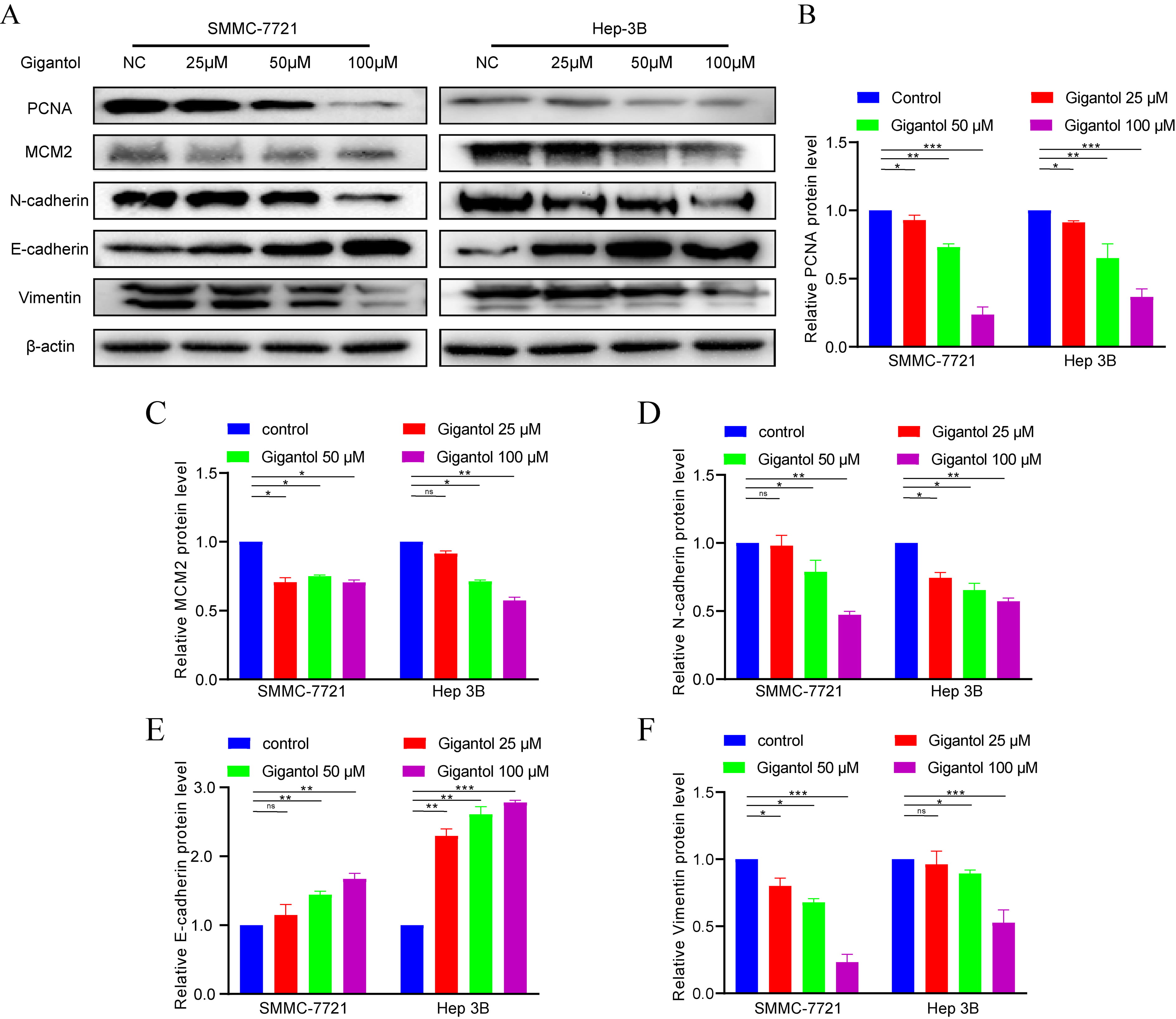 Fig. 8.
Fig. 8.Effects of gigantol on biomarkers of Phenotypes. (A,B,C) (D,E,F) Detection of proliferation and EMT biomarkers using western blot analysis. Data of quantifiable western blot assays.
To verify the regulatory effect of gigantol on target proteins, we examined the changes of protein levels after gigantol intervention. As can be seen from Fig. 9, the expression levels of ESR1, HSP90AA1, and XIAP were gradually depressed with the increase of drug concentration. Combined with the results obtained in Fig. 4, we found that the drug target HSP90 protein is the upstream regulator of Akt. Therefore, we tentatively examined the changes of downstream p-Akt, Akt and CDK1 protein levels. As expected, we found that the levels of p-Akt and CDK1 were subsequently altered after drug intervention. This is consistent with the altered levels of the 3 targets.
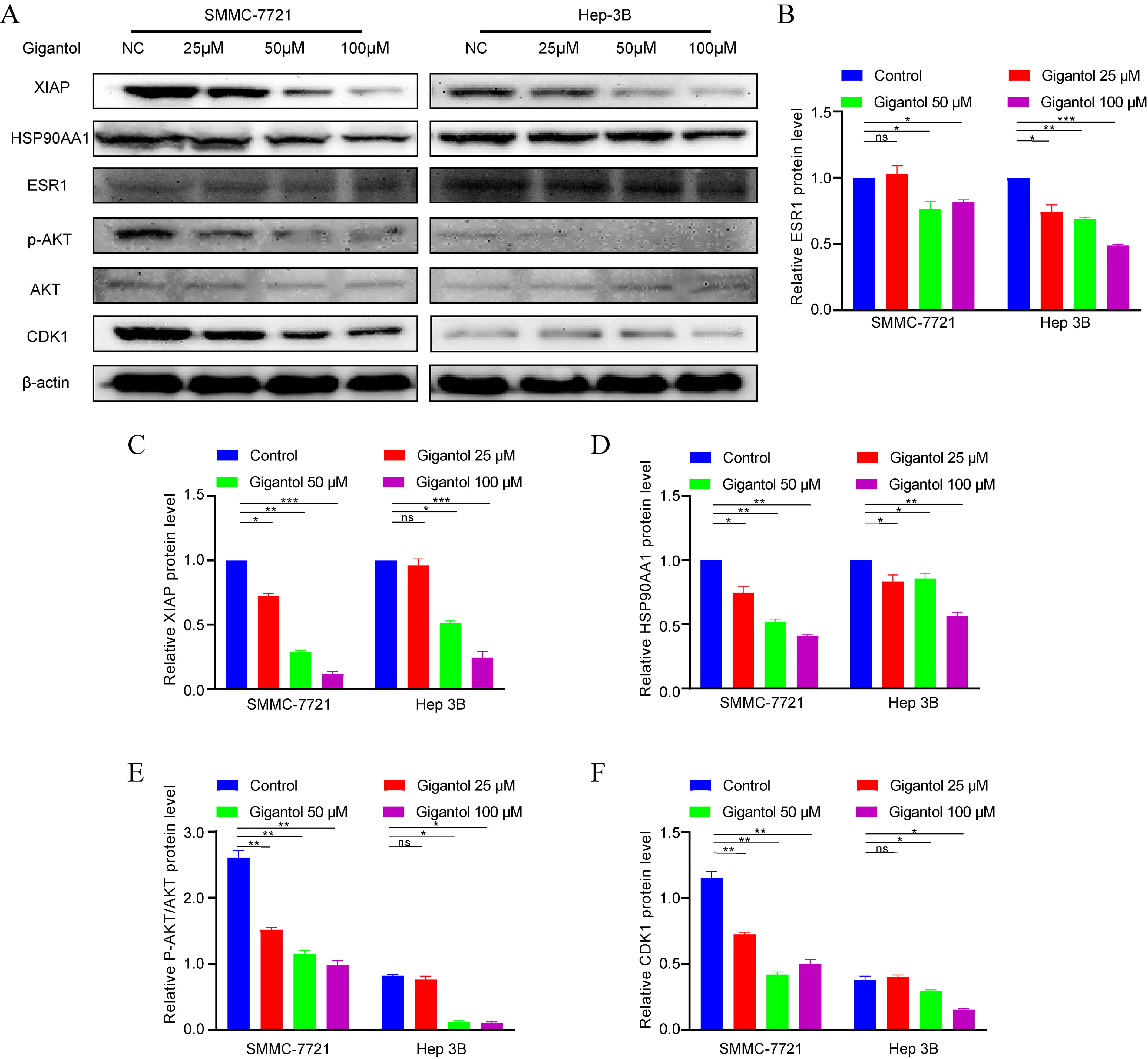 Fig. 9.
Fig. 9.Effects of gigantol on drug targets and HSP90/Akt/CDK1 signaling pathway. (A,B,C,D,E,F) Detection of ESR1, HSP90AA1, XIAP, p-Akt, Akt and CDK1 using western blot analysis. Data of quantifiable western blot assays.
In this study, a “drug-target-disease” network was constructed using network pharmacology to explore the mechanism of gigantol in the treatment of HCC. Gigantol can exert its efficacy in the treatment of HCC through multiple targets and pathways (Fig. 10). The results of network pharmacology analysis demonstrated that there were 32 targets related to gigantol, corresponding to 30 significant signaling pathways, including tumorigenesis, the cell cycle and immune regulation.
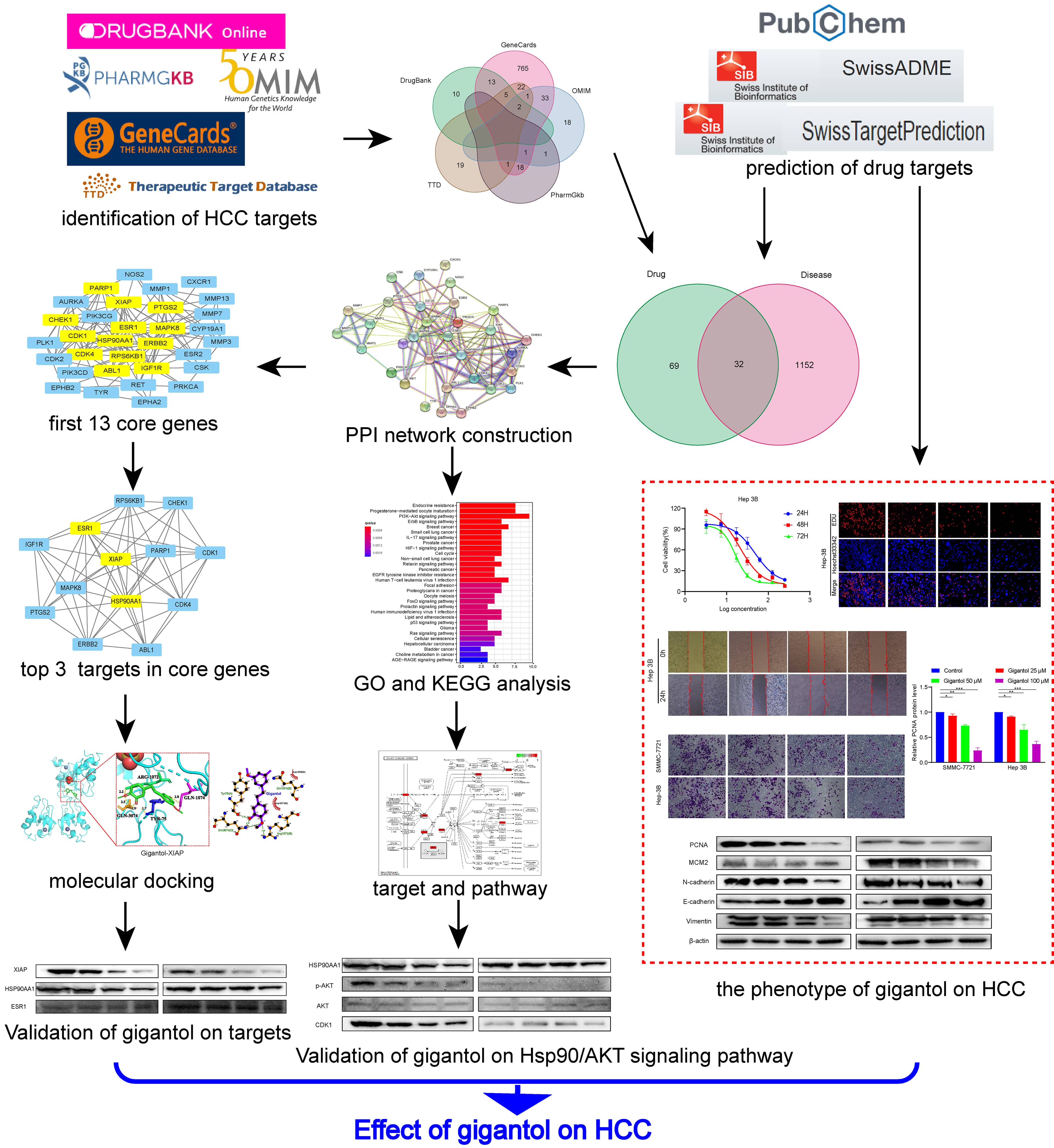 Fig. 10.
Fig. 10.Workflow of network pharmacology analysis of gigantol against HCC. Through database mining, a series of online tools, two categories of targets were identified, including (1) Targets of gigantrol, and (2) genes associated with HCC. The intersection of these two sets of targets may be part of the potential therapeutic targets for hepatocellular carcinoma. Then, PPI, KEGG, and GO analyses were performed for all the obtained targets. In addition, molecular docking was performed to predict the possible targets of action. Finally, the phenotype and HSP90/Akt/CDK1 pathway effects of gigantol on the role of HCC were verified.
The bioavailability of gigantol is high, and it is known from the SwissADME
results that gigantol is easily absorbed through the gastrointestinal tract. In
drug likeness, Lipinski, Ghose, Veber, Egan, and Muegge scores were high. Some
studies reported that gigantol could inhibit the metastasis of human bladder
cancer cells through the Wnt/EMT signaling pathway [37]. One study found that
gigantol could inhibit the proliferation of lung cancer by enhancing the
GSK3-
The results of KEGG pathway analysis showed that signaling pathways, such as the
PI3K/Akt signaling pathway, cancer pathway, IL-17 signaling pathway, and HIF-1
signaling pathway, were significantly different. Among these pathways, the
PI3K/Akt signaling pathway and HIF-1 signaling pathway are closely related to the
occurrence of HCC. The PI3K/Akt signaling pathway is an important pathway for
transmitting extracellular signals to the nucleus. Akt is an important link
between intracellular growth regulation and glucose metabolism and plays an
important regulatory role in various biological activities, such as cell
proliferation, apoptosis, survival and metabolism [41]. Akt is an important
downstream mediator of the PI3K promoter. After phosphorylation activation, PI3K
regulates the expression of various genes, such as those related to apoptosis,
the cell cycle and metabolism, by regulating downstream targets FOXO-1 and GSK-3
[42]. HIF-1
The PPI network showed that the 13 node degree values of ABL1, RPS6KB1, CDK4, ERBB2, HSP90AA1, XIAP, CDK1, ESR1, MAPK8, PARP1, CHEK1, IGF1R and PTGS2 were significantly higher than those of the other targets. Among them, the target proteins ESR1, XIAP and HSP90AA1 appear to be the most relevant and play a crucial role. Abnormal expression of the estrogen receptor 1 gene can promote a variety of liver diseases, such as chronic alcoholic liver disease [46], HBV-related cirrhosis [47], benign liver adenoma [48], and malignant liver cancer, and even affect the therapeutic effect of entecavir [49]. Compared with normal individuals, patients with liver cancer are more likely to have mutations and abnormal expression of the ESR1 gene. ESR1 mutations may be an important marker in the progression of liver disease, and HBV carriers with ESR1 mutations have a higher probability of developing malignant changes [50].
X-linked inhibitor of apoptosis protein (XIAP) is a member of the inhibitor of apoptosis family of proteins (IAPs). XIAP is the most prevalent and potent apoptosis inhibitor protein in the IAP family. XIAP contains a ring-finger structure with ubiquitin ligase (E3) activity. The E3 ligase of XIAP can upregulate cell cycle protein D1 transcription through transactivation of c-Jun/AP-1, which in turn affects the signaling cascade responses of apoptosis, proliferation and migration [51]. In contrast, when XIAP expression was down-regulated, cell cycle protein D1 expression was reduced and cell proliferation was attenuated [52]. In addition, XIAP promotes the proliferation of hepatocellular carcinoma cells by regulating the expression of the CDK4/CDK6/CyclinD1 complex. After treatment of hepatocellular carcinoma cells with embelin (an XIAP-specific inhibitor), tumor cell proliferation was significantly inhibited [53, 54].
The heat shock protein (HSP) family consists of molecular chaperone proteins that play an important role in tumor development. Studies have shown that HSP90 can play a role in inhibiting apoptosis, regulating cell division and promoting angiogenesis [55]. Further studies have revealed that HSP90 is highly expressed in a variety of tumors, such as lung and liver cancers [56, 57]. As an important member of this family, HSP90AA1 has been shown to have pro-proliferative and prognostic effects in tumors such as breast cancers [58]. HSP90 plays a key role in the conformational maturation of oncogenic signaling proteins, among them HER-2/ErbB2, Akt, Raf-1, Bcr-Abl, and mutant P53, as a molecular chaperone [59, 60]. HSP90, has an essential role in the PI3K/Akt/mTOR complex of the PI3K/Akt signaling pathway to promote endothelial cell survival as a key protein chaperone [61, 62]. Our results revealed that PI3K/Akt pathway is one of the key pathways interrupted by gigantol in hepatocellular carcinoma. Further analyzing with the molecular docking results, we found that HSP90 is the protein playing a key role in the PI3K/Akt pathway. We further explored the role of HSP90/Akt and downstream protein CDK1 on this work and validated the results from in vitro experiments.
The molecular docking results indicated that gigantol could bind stably to the core target proteins ESR1, XIAP and HSP90AA1, indicating that gigantol may regulate cell proliferation and apoptosis by acting on these targets to modulate immune activity and thus treat HCC. The same conclusion can be tentatively drawn from our western blotting validation results. Among them, gigantol has the strongest ability to regulate XIAP protein. In addition, the results from in vitro experiments showed that gigantol inhibited the growth, invasion, and metastasis of hepatocellular carcinoma cells in a dose-dependent manner. Additionally, we detected the expression of related marker proteins using western blotting, all of which yielded similar results. We verified from molecular docking and protein level that gigantol can exert regulatory effects through three targets, ESR1, XIAP and HSP90AA1, but it is never enough. In the future, it is worthwhile to further resolve the structure by cryoelectron microscopy or X-ray diffraction crystals.
In conclusion, gigantol has multiple effects, such as tumor regulation and immune modulation, and is mainly used to treat HCC by regulating the expression of related proteins through the multiple above mentioned components, targets and pathways. However, Chinese medicine is a complex multicomponent system. The present study only confirmed the mechanism of gigantol in the treatment of liver cancer from a microscopic perspective. Nevertheless, the study lacks data for in vivo experimental validation. Follow-up studies will be conducted based on the present study to evaluate gigantol’s clinical application.
Our findings suggest that gigantol can treat hepatocellular carcinoma through mechanisms predicted by network pharmacology, such as inhibition of Hep3B and SMMC-7721 cell proliferation, invasion, metastasis and expression of related proteins. These findings provide potential molecular and cellular evidence that gigantol may be a promising antitumor agent. In addition, we preliminarily confirmed three key drug targets of gigantol, which are ESR1, HSP90AA1 and XIAP. Further explored that gigantol may exert its effects through the HSP90/Akt/CDK1 signaling pathway.
HCC, Hepatocellular carcinoma; RFA, Radiofrequency ablation; TACE, Transarterial chemoembolization; Akt, Protein kinase B; OCT4, Organic cation/carnitine transporter 4; Nanog, Nanog homeobox; BBB, Blood brain barrier; GO, Gene ontology; BP, Biological process; CC, Cellular component; MF, Molecular function; KEGG, Kyoto encyclopedia of genes and genomes; PI3K, Phosphatidylinositol-4,5-Bisphosphate 3-Kinase; ErbB, Epidermal growth factor receptor (EGFR); ESR1, Estrogen receptor 1; HSP90AA1, Heat shock protein 90 alpha family class a member 1; XIAP, X-linked inhibitor of apoptosis; PCNA, Proliferating cell nuclear antigen; MCM2, Minichromosome maintenance complex component 2; CDK1, Cyclin dependent kinase 1.
SL, HL—conceptualization, methodology, visualization, data curation, writing original draft; SL, DY, XX, XL, XC—software, data curation; JL, XX, HL—validation, writing, review, editing; YY—supervision, project administration; All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
Not applicable.
We sincerely appreciate the technical support from Nanjing University of Chinese Medicine.
This study was funded by the Fujian Province Key Specialties in Traditional Chinese Medicine Construction Project (No. Fujian Health Chinese Medicine Letter (2019) No.262), the Jiangsu Province condition construction and livelihood science technologic special funds (BL2014005), the Startup Fund for scientific research, Fujian Medical University (Grant number: 2020QH1080), the 2021 Jiangsu Postgraduate Research and Practice Innovation Program of Postgraduate Training Innovation Project (SJCX21_0761), and Medical Science and Technology Development Foundation, Nanjing Department of Health (YKK19112).
The authors declare no conflict of interest.
