1. Introduction
Acute lymphoblastic leukemia (ALL), in particular B-cell precursor ALL, is the
most frequent hematological malignant neoplasm occurring during childhood, with a
peak incidence between 1 and 4 years of age. The standard ALL therapy lasts 24
months and therapeutic protocols comprise different chemotherapeutic and
immunosuppressive agents [1]. Although often burdened by severe side effects,
fist-line polychemotherapeutic approaches result in clinical remission in most
pediatric patients (5-year survival rate approximately 80%) [2, 3, 4]. In contrast,
ALL could be fatal in ~50–60% of children with recurrence [5].
In relapsed/refractory ALL, haematopoietic stem-cell transplantation might
represent the only curative option; however, this clinical procedure requires
bone marrow remission that is not easily achievable in patients who already
failed to respond to pharmacological treatments [6]. A relevant ongoing approach
in ALL therapeutic protocols is the introduction of new biological drugs, which
should be integrated in salvage therapy to further improve rescue outcome. Among
these, there is the bispecific T-cell engager (BiTE) antibody construct
blinatumomab (Blincyto) [7]. Blinatumomab is a single peptide with a
molecular weight of ~55 kDa and consists of two antibody single
chain variable fragments (one against the surface antigen CD3 and one against
CD19) joined by a flexible glycine-serine linker. Blinatumomab acts as a short
adapter molecule that forces mature CD3 cells (T-lymphocytes) and
CD19 tumor B cells into close proximity, creating a tight cell-specific
synapse that induces a strong T-cell activation and proliferation. Effector cells
(CD3) are thus able to recognize leukemic blasts and induce their death by
apoptosis, through the release of perforins and granzymes [8]. Blinatumomab has
been administered with success in relapsed/refractory B-ALL adult cases. However,
primary resistance has been observed in 20–30% of adult patients [6]. Recently,
two international randomized clinical trials (www.clinicaltrials.gov;
identification numbers: NCT02101853 and NCT02393859) clearly demonstrated the
benefit of blinatumomab in terms of overall survival in pediatric patients with
high-risk first relapse of B-ALL with chemotherapy-responsive disease.
Blinatumomab was administered in combination with conventional chemotherapy and
compared to conventional chemotherapy alone. Importantly, blinatumomab showed a
favorable toxicity profile, with less life-threatening complications [9, 10].
The role of CD19 in therapy failure with blinatumomab has been hypothesized and
investigated to clarify whether blinatumomab exposure exerts a potent selective
pressure, resulting in a CD19 negative recurrence; controversial conclusions on
this issue were reported [11, 12]. CD19 is a co-stimulatory type I transmembrane
protein (~95 kDa), exclusively expressed on B-cells. It functions
as a critical co-receptor of the B-Cell Antigen-Receptor (BCR) signal
transduction pathway. In particular, CD19 mediates the activation of the Src
family protein-tyrosine kinases, such as Lyn and Fyn [13], that enhance
BCR-induced signaling through recruitment and activation of PI3K and downstream
Akt kinases, thus promoting B-cells survival and cell growth [14]. The
CD19 gene contains a binding site for the B-cell lineage specific
activator protein (BSAP) in its promoter’s region, and the expression of CD19 is
reduced in pre-B-cells deficient in BSAP [15]. BSAP is encoded by the
PAX5 gene and is the only PAX protein expressed in the hematopoietic
system. The protein functions both as a transcriptional activator and as a
repressor on different target genes involved in B lymphoid lineage development
[16, 17]. Interestingly, genomic and transcriptomic analyses in pediatric and
adult B-ALL identified PAX5 as one of the most often mutated genes in
leukemic cells, being involved in several leukemia-associated rearrangements,
such as gene fusions (2–3% of ALL cases) or intragenic amplifications [18, 19, 20].
Moreover, several somatic point mutations at the level of the PAX5
exonic sequence, influencing the different structural domains and functions of
BSAP, have been identified in over 30% of ALL cases [19, 21]. The genetic basis
of interpatient variability in blinatumomab response have not been investigated
so far. The aim of this study was thus to provide evidences of the hypothesis
that a differential CD19 surface density on B-ALL cells, due to a heterogeneous
PAX5 genetic background, could affect blinatumomab response. Therefore,
co-cultures of T-lymphocytes from healthy donors and lymphoblastic B-ALL cell
lines (with a different PAX5 genetic status) were maintained in the
presence of blinatumomab for one week. Cytofluorimetric analyses investigating
cell morphology, CD19CD3 composition and viability were performed
after 3 and 7 days of incubation with the drug. Similar in vitro
analyses were performed on primary cells co-cultures derived from bone marrow
aspirates of ALL patients.
2. Materials and methods
2.1 Reagents
Blinatumomab (Blincyto®, Amgen, Italy) was provided as a sterile
12.8 g/mL infusion solution and kept at –20 C until use.
Recombinant human interleukin-2 (IL-2, I2644, Sigma-Aldrich, Italy) was suspended
at a final concentration of 10 g/mL in sterile phosphate-buffered saline
(PBS, D8537, Sigma-Aldrich, Italy), supplemented with 0.1% human serum albumin.
2.2 Cell cultures
Human immortalized CD19 leukemia cell lines, NALM6 (ACC-128) and REH
(ACC-22), were purchased from DSMZ (Leipzig, Germany). Cell cultures, trypan blue
exclusion assay, lysates preparation and western blot analyses are described in
supplementary materials.
2.3 Study populations
Buffy coats of three healthy blood donors were provided by the
Transfusion Service of the “Azienda Sanitaria Universitaria Giuliano Isontina”
(ASUGI, Trieste, Italy). Patients were ALL children enrolled at IRCCS Burlo
Garofolo in Trieste in the AIEOP-BFM ALL 2009 protocol. According to the AIEOP
ALL protocol guidelines, karyotyping and cytogenetics were performed as part of
the diagnostic procedures. The clinical study was approved by the local ethical
committee (Protocol number CE/V 135; IRCCS Burlo Garofolo, March 5th 2012) and
appropriate informed consent was obtained from patients/donors and/or their
parents or guardians.
Blood samples were processed for the isolation of peripheral blood mononuclear
cells (PBMC) and purification of primary T-lymphocytes (CD3) and stored at
–80 C, as described in Supplementary material.
2.4 Cytofluorimetric assay
T-lymphocytes (CD3) isolated from 3 healthy donors were thawed and seeded
in a 24-wells plate at 1 10 cells/ml in complete RPMI-1640
medium supplemented with IL-2 at 10 ng/mL, to allow their recovery. After 24
hours (day 0), co-cultures of 5000 T-lymphocytes/well and 50,000 B-ALL cell lines
(NALM6 or REH)/well were set up (seeding effector-to-target ratio 1:10, 200
L in X-VIVO 15 plus 10 ng/mL IL-2 and blinatumomab (0, 1, 10
ng/mL). Plates were incubated at 37 C (5% CO) for 7 days; every
24 hours, 150 L of the supernatants were replaced by fresh complete
culture medium blinatumomab [22]. Removed supernatants were stored at
–20 C for subsequent granzyme B (GzB) quantification. Cytofluorimetric
analyses were performed on day +3 and day +7, using 10 colors/3 lasers Navios EX
(Beckman Coulter, CA, USA), acquiring at least 30,000 events for each sample.
Briefly, 100 L of the cell suspensions were stained for surface antigens
(CD45, CD19, CD3) and for intracytoplasmic markers (DNA and nuclei) using 20
L of a cocktail of fluorescent dyes (anti-human antibodies: antiCD45-KO,
antiCD19-ECD, antiCD3-PE; 7-amino-actinomycin D (7AAD), Syto16-FITC (all from
Beckman Coulter, Pasadena, CA, USA)). For each antigen, mean fluorescence
intensity (MFI) was calculated using the Kaluza Analysis Software version 2.1
(Beckman Coulter, CA, USA).
The cytofluorimetric assay was also performed on patients’ mononuclear cells.
Due to the scarce biological material available, experiments could be performed
only once per patient. Six hundred thousand cells were seeded in triplicate in
X-VIVO 15 medium IL-2 (10 ng/mL) blinatumomab (0, 1, 10
ng/mL) for 7 days. Every 24 hours, supernatants were refreshed. Cytofluorimetric
analyses were performed at day +7, according to the conditions described above.
2.5 GzB ELISA assay
The human GzB Platinum ELISA kit (BMS2027, Thermo Fisher Scientific, Italy) was
used according to manufacturer’s instructions.
2.6 Statistical analysis
For the cytofluorimetric assays on co-cultures including immortalized B-ALL cell
lines, three independent experiments were performed in triplicate and data are
presented as mean SD; for each patient sample, the assay was performed
once, in triplicate. Data were compared by two-way ANOVA followed by Bonferroni
post-test. For the GzB ELISA assay, three-way ANOVA followed by Bonferroni
post-test was used. Data were analyzed by GraphPad Prism software version 8.3.0
(CA, USA) and significant differences were considered at p values
0.05.
3. Results
3.1 NALM6 and REH cells showed a differential expression of PAX5 and
CD19
According to the literature, both NALM6 and REH cell lines harbor complex
karyotypes and rearrangements of chromosome 12, leading to the expression of ETV6
chimeric fusion oncoproteins (ETV6-PDGFRB in NALM6 [23] and ETV6-RUNX1 in REH
[24]). Moreover, NALM6 cells harbor a focal, heterozygous deletion in the
PAX5 promoter [25] and a homozygote point mutation in an intronic,
non-coding poly(C) microsatellite repeat, which is unlikely to be functional
[26]. REH cells instead carry a PAX5 frameshift mutation, due to a
single base insertion (C) within the coding poly(C)7 microsatellite repeat in
exon 8 (InsP321fs). This results in premature termination of translation after
amino acid 339, encoding a BSAP protein lacking the C-terminal transactivation
domain [26]. To explore the relevance of these PAX5 genetic alterations
in blinatumomab response, NALM6 and REH cells were employed as representative
in vitro models. Differential expression of BSAP and CD19 in the two
cell lines was confirmed at the protein level (Fig. 1A and Supplementary
Fig. 1). Western blot results, using a commercial antibody directed against the
N-terminal part of human BSAP, detected the full-length protein (42 kDa) in
NALM6, clearly expressed if compared to REH where only a faint signal appeared;
no additional truncated form of BSAP was visible (Fig. 1A). The CD19 protein
surface expression on both NALM6 and REH cells was investigated by flow
cytometry, staining 1 10 cells with the anti-CD19-ECD antibody.
Both cell lines, being CD19, showed a fluorescence peak caused by the
direct binding of the antibody to the antigen; however, a forward shift in the
CD19 peak with a three times higher fluorescence intensity (median: 28895
versus 9155) in NALM6 compared to REH was observed, suggesting a higher
surface density of the antigen in the former cells (Fig. 1B). PAX5 and
CD19 mRNA expression levels were comparable among the two cell lines, as
shown by SYBR Green qPCR (Supplementary Fig. 2).
 Fig. 1.
Fig. 1.
PAX5 and CD19 protein expression in B-ALL cell lines, NALM6 and
REH. (A) Detection of PAX5 expression in REH and NALM6 cells by western
blotting; Vinculin blot used as loading control. (B) Flow cytometry analysis of
CD19 expression on cell surface; 1 10 live cells were directly
labelled with an anti-CD19-ECD antibody for 20 min at room temperature in the
dark. X-axis represents the fluorescence intensity.
3.2 Co-culture of NALM6 or REH cells and T-lymphocytes respond
differently to blinatumomab in vitro
Preliminary analyses were carried out to evaluate cell survival of immortalized
CD19 B-ALL cell lines and CD3 T lymphocytes in X-VIVO 15
cultural medium without blinatumomab. The number of cells was monitored daily
over a 7 day-period by trypan blue exclusion assay (initial seeding
concentration: CD3 lymphocytes: 84,000 cell/mL; CD19 cells:
62,500–500,000 cells/mL). NALM6 or REH cells could proliferate in this
serum-free medium: a seeding concentration of 250,000 B-cells/ml in 96-wells
plates (50,000 cells/well) was chosen to guarantee a similar number of alive
growing NALM6 or REH cells over one week in the untreated condition
(Supplementary materials, Supplementary Fig. 3). In contrast,
cell viability of T-lymphocytes was severely impaired from the first day of
incubation, and survival was not increased by the addition of IL-2 in the culture
medium (Supplementary Fig. 4).
To test whether the increased surface presentation of CD19 in B-cells affects
blinatumomab in vitro response, co-cultures of CD3 isolated
lymphocytes of three different healthy donors and either NALM6 or REH cells were
set up at a 1:10 ratio (initial 5000 T-cells: 50,000 B-cells). X-VIVO 15
medium supplemented with IL-2 and fresh blinatumomab (when required) was replaced
every 24 hours. Fig. 2 shows exemplificative results of the cytofluorimetric
analysis performed after 3 and 7 days of incubation, respectively. Fig. 3 reports
the mean percentage value ( SD) of B-ALL cells and T-lymphocytes, as
measured by the SS/CD45 analysis. After 3 days of incubation, there was no
difference between the condition with or without blinatumomab in the co-cultured
B- and T-cells (Fig. 3A). Changes were on the contrary observed after 7 days
(Fig. 3B), with a significant decrease in the B-ALL cell percentage in treated
compared to untreated samples (two-way ANOVA, Bonferroni post-test, p 0.0001); the decrease was particularly evident in REH compared to NALM6 after
drug exposure (ANOVA two-way, Bonferroni post-test, p 0.05). At the
same time point, treatment with blinatumomab 1 ng/mL significantly increased
T-lymphocytes in comparison to controls (two-way ANOVA, Bonferroni post-test,
p 0.0001), with a more important effect for T-lymphocytes in
co-culture with REH than with NALM6 (two-way ANOVA, Bonferroni post-test,
p 0.05).
 Fig. 2.
Fig. 2.
Exemplificative flow cytofluorimetric analysis of the CD3
cells:CD19 cells co-culture (1:10) after 3 days (upper square panel) and 7
days (lower square panel) of incubation in the absence (panels A–C) or presence
of blinatumomab 1 ng/mL (panels D–F). SS/FS panels (panels A and D) and CD45/SS
panels (panels B and E) are commonly acquired during diagnostic procedure in ALL,
and combine cell morphological information of size and intracellular complexity
(FS, SS) with labeling of a pan-leukocyte marker (CD45), distinguishing between
immature lymphoblasts (larger, with a variable but moderate staining of CD45)
from mature lymphocytes (smaller, with high CD45 and a near-total absence of
intracellular complexity). 7-AAD/Cyto16 labelling (panels C and F) assayed cell
viability. B-ALL cells are highlighted in red, healthy donors T-cells in blue.
 Fig. 3.
Fig. 3.
Mean percentage composition of T-lymphocytes and B-ALL cell
lines in the co-culture (initial effector-to-target seeding ratio, 1:10) after 3
(A) and 7 (B) days of incubation in the absence or presence of blinatumomab 1
ng/mL. Error bars represent SD (n = 3). * p-value 0.05, ****
p-value 0.0001, two-way ANOVA, Bonferroni post-test. After 3 days of incubation no difference was observed in co-cultures treated with or without blinatumomab (Panel A); after 7 days of incubation (Panel B), a significant decrease in the B-ALL cell percentage and an increase in T-lymphocytes percentage in samples treated with blinatumomab was observed.
7AAD and Syto16 labelling were used to analyze cell viability; Fig. 4 reports
the percentage value of dead NALM6 or REH (CD19, 7AAD) and of alive
co-cultured T-cells (CD3, 7AAD/Syto 16), measured in flow
cytometry. A significant increase in mortality of both leukemic cell lines was
observed after incubation with blinatumomab 1 ng/mL in comparison to untreated
cells at both time points (untreated versus treated, day +3, p-value
0.001, Fig. 4A; day +7, p-value 0.0001, two-way ANOVA, Bonferroni
post-test, Fig. 4B). Mortality in cells treated with blinatumomab was higher in
REH compared to NALM6 at day +3 (44.46 6.9% versus 25.54 12.9%,
respectively, two-way ANOVA, Bonferroni post-test, p 0.05, Fig. 4A)
and was almost complete in both cell lines at day +7 (REH: 95.12
2.1% NALM6: 97.17 1.8%). Statistical analyses did
not show any difference in T-lymphocytes survival neither comparing those exposed
to blinatumomab to the unexposed ones at both 3 and 7 days (percentage of alive
cells 80%), nor comparing T-lymphocytes co-cultured with NALM6 to those
co-cultured with REH cells (Fig. 4).
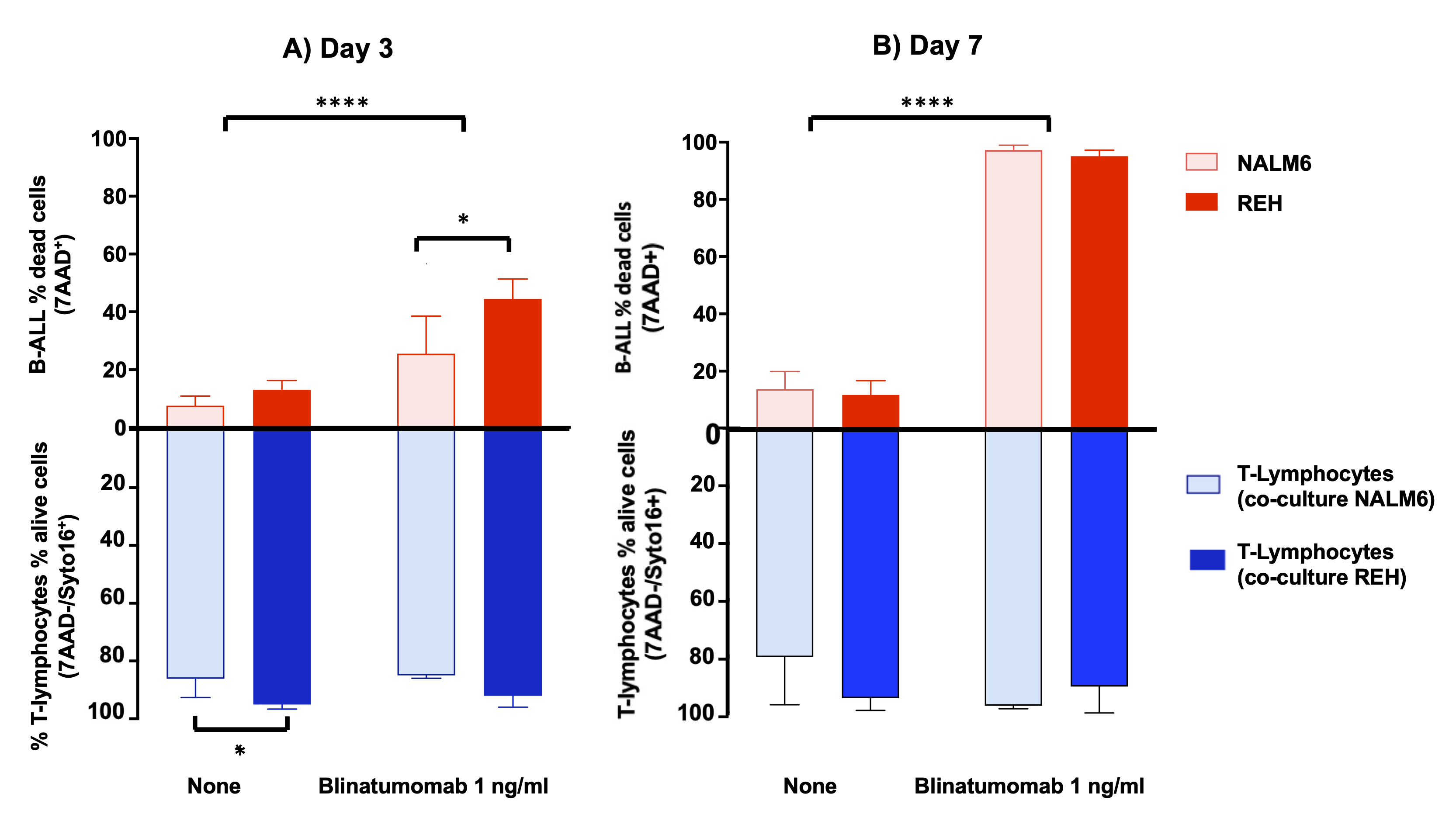 Fig. 4.
Fig. 4.
Mean percentage value of T-lymphocytes viability and B-ALL cell
lines mortality in the co-culture (initial effector-to-target seeding ratio,
1:10) after 3 (A) and (7) days of incubation in the absence or presence of blinatumomab 1 ng/mL. Error bars represent
SD (n = 3). * p-value 0.05, *** p-value 0.001, ****
p-value 0.0001 two-way ANOVA, Bonferroni post-test. A significant increase in mortality of both leukemic cell lines is observed after incubation with blinatumomab 1 ng/mL in comparison to untreated cells at both time points. Mortality in cells treated with blinatumomab is higher in REH compared to NALM6 at day +3 (Panel A) and is almost complete in both cell lines at day +7 (Panel B). No effect on T-lymphocytes viability was observed.
3.3 Co-culture of NALM6 or REH and T-lymphocytes show a different
blinatumomab induced GzB daily release over time
GzB is a mammalian aspartic acid-cleaving serine protease, released by cytotoxic
T lymphocytes. GzB was measured by an ELISA assay in supernatants collected every
24 hours, as a measure of daily T-lymphocytes activation. Data are shown in Table 1 and Supplementary Fig. 5. In the untreated condition, GzB levels were
low, although constantly growing over time, likely because of an intrinsic
drug-independent mechanism of T-lymphocyte activation; levels were comparable
over time between co-cultures including either NALM6 or REH. Blinatumomab
enhanced the daily release of GzB compared to untreated controls at any time
point and regardless of the B-ALL cell line present in the co-culture
(p-value 0.0001, three-way ANOVA). Concentrations of GzB reached the
maximum (3478.1 pg/mL) already at day +3 in NALM6 co-culture, and were
significantly lower in REH co-cultures at days +3 and +4 (p-value 0.0001, three-way ANOVA), reaching a comparable plateau only at day +5.
In REH co-cultures, the 24-hours release of GzB was almost doubling between days
+3 and +4 and days +4 and +5.
Table 1.GzB daily release in CD3:CD19 co-cultures (initial
effector-to-target seeding ratio, 1:10).
|
NALM6 co-culture |
REH co-culture |
|
| GzB (pg/mL) |
GzB (pg/mL) |
|
| Days |
Untreated |
Blinatumomab |
p* |
Untreated |
Blinatumomab |
p* |
p |
p‡ |
| 1 ng/mL |
1 ng/mL |
| +3 |
37.2 19.2 |
3478.1 |
0.0001 |
9.4 4.6 |
1073.0 29.5 |
0.0001 |
Ns |
0.0001 |
| +4 |
100.6 66.5 |
3478.1 |
0.0001 |
28.2 11.2 |
2091 216.0 |
0.0001 |
Ns |
0.0001 |
| +5 |
249.5 149.4 |
3478.1 |
0.0001 |
205.6 111.5 |
3478.1 |
0.0001 |
Ns |
Ns |
| +6 |
288.1 295.6 |
3478.1 |
0.0001 |
725.2 636.4 |
3478.1 |
0.0001 |
Ns |
Ns |
| +7 |
648.5 376.1 |
3478.1 |
0.0001 |
696.7 201.7 |
3478.1 |
0.0001 |
Ns |
Ns |
| Mean values SD (n = 3) are reported. p-value calculated
according to three-way ANOVA: * basal (untreated) versus blinatumomab (1
ng/mL) GzB induced release in NALM6 and REH co-cultures, † basal GzB
release comparison between NALM6 and REH co-cultures, ‡
blinatumomab (1 ng/mL) GzB induced release comparison between NALM6 and REH
co-cultures. GzB, granzyme-B; Ns, not significant. |
3.4 Patient primary co-cultures of B-ALL lymphoblasts and
T-lymphocytes respond differently to blinatumomab in vitro
Blinatumomab in vitro assay was performed on primary mononuclear cells
co-cultures of 8 patients with Philadelphia-negative B-ALL; results are available
only for 5 children (median (IQ) age: 10.7 (7.9–11.3) years; males: 60%): in
the other 3, the assay could not be concluded due to technical reasons. CD3lymphocytes and primary CD19 leukemic cells were derived from bone
marrow aspirates collected at diagnosis: percentages of immature CD19
B-cells exceeded 66% in the diagnostic samples (mean SD: 84.40
11.55%) whereas mature CD19 B-lymphocytes and CD3 T-lymphocytes were
1.15 1.04% and 4.65 3.84%, respectively; the CD3:
CD19 cells ratio was at least equal to 1:8 (Table 2). Blasts PAX5
genetic status was not known.
Table 2.Patients’ demographic characteristics and CD3/CD19 cells in the diagnostic bone marrow.
| Patients |
Age |
Gender |
Diagnosis |
Genetic alterations |
Immunophenotype |
CD19 immature cells (%) |
Mature cells |
Effector-to target ratio |
| CD3 (%) |
CD19 (%) |
| #1 |
14.6 |
M |
2nd RELAPSE |
None |
ALL-B COMMON |
66 |
6.2 |
2.8 |
1:11 |
| #2 |
7.9 |
M |
1st ONSET |
MLL/AF4 |
ALL-B COMMON |
86 |
3 |
0.4 |
1:30 |
| #3 |
7.0 |
F |
1st ONSET |
None |
ALL-B COMMON |
82 |
10.7 |
1.6 |
1:8 |
| #4 |
10.7 |
F |
1st ONSET |
None |
NA |
95 |
1.4 |
0.3 |
1:68 |
| #5 |
11.3 |
M |
1st ONSET |
None |
ALL pre-B |
93 |
2 |
0.7 |
1:47 |
Fig. 5 reports the mean percentage value ( SD) of blast cytotoxicity and
T-lymphocytes activation after 7 days of in vitro incubation, measured
by the CD19 gated 7AAD and by the CD3 gated
SS/CD45 cytofluorimetric panels, respectively. Under basal condition, blasts
cytotoxicity was highly variable among samples (two-way ANOVA, Bonferroni
post-test, p 0.001 for untreated CD19 cells) and was not
affected by the addition of IL-2 in the cultural medium. In contrast, comparable
levels of alive T-lymphocytes were observed and were slightly enhanced by the
presence of the cytokine (CD3 cells: mean percentage value ( SD):
2.97 (1.11–3.27)% and 1.80 (1.74–2.60)% with and without IL-2 respectively;
p 0.0001). A positive in vitro pharmacological response to
blinatumomab was defined as a simultaneous significant increase in blast
mortality and T-lymphocytes activation induced by the drug, whereas
non-simultaneous or failed enhancements were considered as resistance. In the
IL-2 free condition, only two samples responded properly to blinatumomab
(p 0.0001 for both CD19 and CD3 cells), although at
different drug concentrations (pt#1 at 10 ng/mL, pt#2 at 1 ng/mL). Blinatumomab
failed to induce any effect in samples derived from pt#3, pt#4 and pt#5. The
presence of IL-2 in the cultural medium induced a significant increase in
T-lymphocytes compared to basal condition in four out of five patients (all but
pt#5). The lower concentration of blinatumomab was thus sufficient to induce a
response in pt#1 (p 0.0001 for both CD19 and CD3
cells). In pt#3, CD3 cells switched significantly from 3.27% to 17.42%
in drug-free and blinatumomab-treated samples (p 0.0001)
and in pt#4 from 0.41% to 10.03% (p 0.0001), however, in both
cases without effect on blasts mortality (pt#3: from 8.02% to 14.16%,
p 0.05; pt#4: from 5.05% to 9.07% respectively, p
0.05). Interestingly, pt#5 remained resistant and did not show any change
compared to IL-2 free condition. These in vitro results are unrelated to
the percentages of immature CD19 B-cells present in the diagnostic samples.
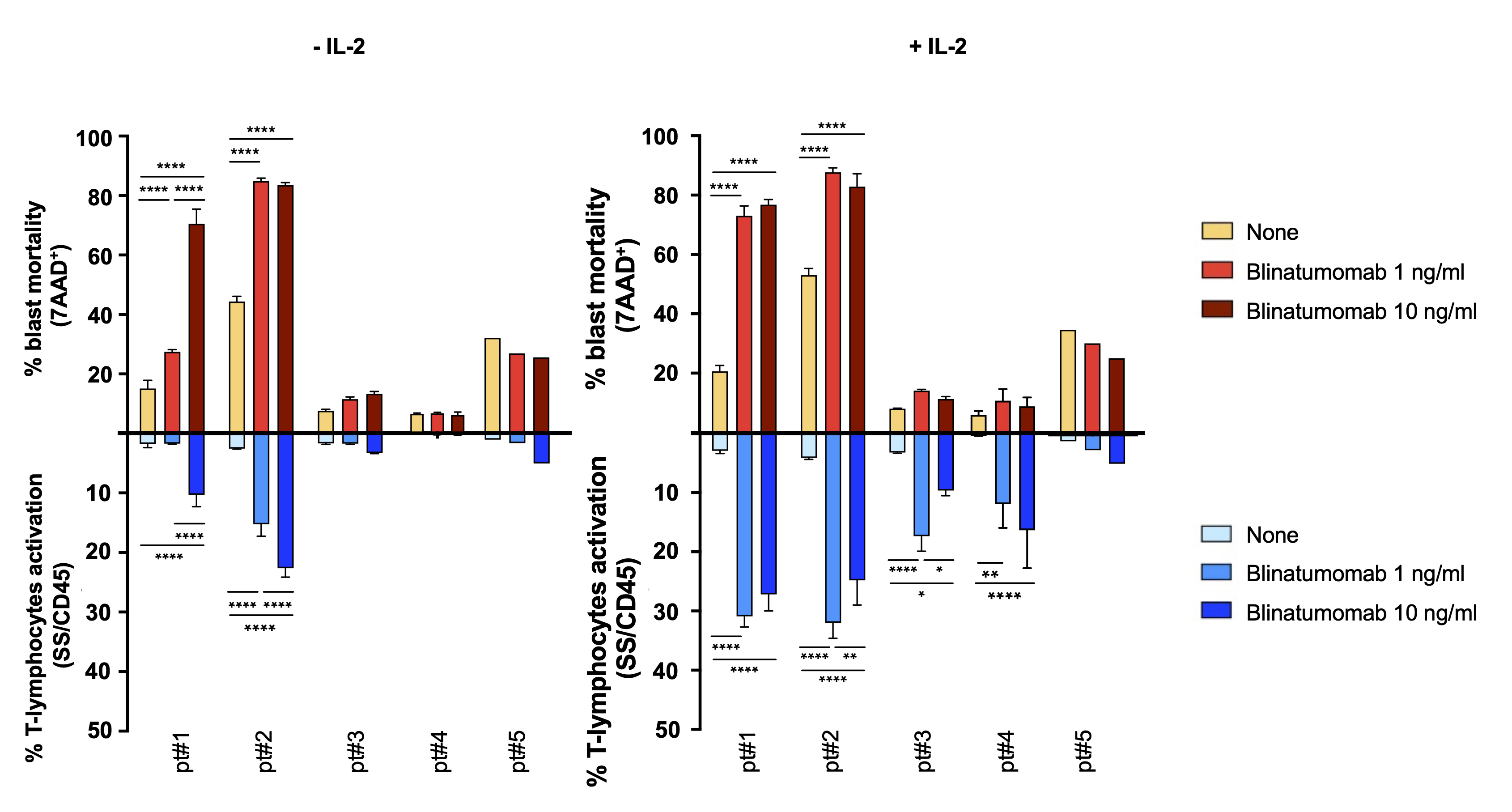 Fig. 5.
Fig. 5.
Blinatumomab in vitro cytofluorimetric assay on
patients’ samples. Mean percentage value (+SD) of patients blasts mortality
(gate CD19, 7AAD, in red) and T-lymphocytes activation (gate
CD3, SS/CD45, in blue) after 7 days of incubation (initial seeding 600,000
cells/well). * p-value 0.05, ** p-value 0.01, ****,
p-value 0.0001, two-way ANOVA, Bonferroni post-test.
4. Discussion
Blinatumomab is a promising agent for both first-onset and relapsed/refractory
ALL, however, primary and secondary resistance occurs, and still needs to be
characterized. Current recruiting clinical trials investigate on blinatumomab
effectiveness in comparison to standard therapeutic procedures (e.g., AIEOP-BFM
ALL 2017 protocol, NCT03643276) and/or on the proportion of patients with
blinatumomab poor-response as primary outcomes. Pharmacogenetic traits or
B-lymphoblasts genetics could modulate treatment efficacy or predispose to
treatment failure. Only few reports on a very limited number of patients suggest
that blinatumomab could be a valid therapeutic approach for specific ALL genetic
subtypes. Zhao and coworkers [27] showed that the activation of JAK-STAT
signaling in leukemic cells of 44 patients harboring high-risk CRLF2 and
EPOR-rearrangements is an important determinant of blinatumomab
response. Mouttet and collaborators [28] reported durable remissions after
blinatumomab treatment in 9 patients with a rare subtype of ALL with a dismal
outcome, characterized by high and homogeneous expression of CD19 on blast cells
and by the presence of the t(17;19) (q21-q22;p13), leading to the
TCF3-HLF fusion gene. Moreover, Zhao and collaborators [29] observed, in
44 adults with relapsed or refractory B-ALL treated with blinatumomab, that
multiple mechanisms may contribute to CD19 loss and relapse, including
CD19 mutations, CD19 mutant allele-specific expression,
CD19 low RNA expression and also mutations in the member of the CD19
signaling complex CD81. This underlines the importance of possible variations in
CD19, including PAX5, in relation to treatment response [29]. Identifying
patients’ prognostic biomarkers and/or finding a reliable predictive in
vitro sensitivity assay could be crucial for an effective proactive management
of blinatumomab, especially considering the high costs, the length of treatment
(2 cycles of 28 days continuous infusion), its toxicity [30] and current
percentages of failure (20–30% patients with relapse/refractory ALL) associated
with blinatumomab therapy. Another CD19-targeted immunotherapy, patient-specific
chimeric antigen receptor T cell therapy (CAR-T), was recently introduced in the
clinics with encouraging and impressive response rates in the cure of
relapsed/refractory ALL and other hematological diseases. The choice between
CAR-T and BiTE immunotherapy depends on several clinical features that may affect
efficacy, including relative disease burden, antigen expression, and T-cell
function, as well as patient and disease characteristics. Adverse effects of
CD19-targeted immunotherapies, such as cytokine release syndrome and
neurotoxicity, still need to be reduced: these events may be more frequent and
severe in patients receiving CAR-T [31, 32]. Blinatumomab has the advantage to
use the same standard drug for every person treated and therapy-associated costs
may be lower. Ongoing attempts in CAR-T aim to develop off-the-shelf CAR T-cell
therapies that should be immediately available for use without being manufactured
for each patient. First attempts used CRISPR/Cas9-engineered universal CD19/CD22
CAR-T cells to avoid host immune-mediated rejection when infused in patients,
showing a manageable safety profile and prominent antileukemic activity [33].
In this study, a cytofluorimetric approach was used to measure in vitro
blinatumomab effects on co-cultures of immortalized CD19 B-ALL cells and
primary CD3 T-lymphocytes, to investigate whether the drug response could
be affected by the CD19 surface expression levels determined by the PAX5
genetic status of leukemic cells. The different expression of BSAP and CD19 in
the immortalized cell lines NALM6 and REH was confirmed at the protein level.
Western blot analysis revealed the presence of the full-length BSAP in NALM6,
whereas only a faint signal appeared in REH cells. Analysis of mRNA expression
levels confirmed previous data of Best and co-workers, who showed comparable
PAX5 mRNA levels between these two cell lines [26]. Thus the lower BSAP
levels could be ascribable to the PAX5 frameshift (fs) somatic mutation
(P321fs) harbored by REH, affecting mRNA translation or protein stability.
PAX5 fs mutations occurred in ~2% of ALL patients, as
demonstrated by Gu and co-workers using integrated genomic analysis of 1,988
childhood and adult cases; ~40% of the PAX5 fs
mutations observed lead to truncated BSAP proteins, similar to what is observed
in REH cells, thus confirming this cell line as a useful cellular model [25]. The
lack of the C-terminal transactivation domain functionally impaired BSAP, and the
truncated protein failed to fully-activate BSAP-dependent gene transcription [34, 35]. Indeed, a three times lower mean fluorescence intensity for CD19 was
observed in REH compared to NALM6 cells, as previously reported by Haso and
collaborators [36]. Indeed, these authors calculated the surface antigen density
on different B-ALL cell lines, finding that the average number of CD19 molecules
per cell in REH was 15,000 versus 55,000 in NALM6 [36]. Preliminary analyses were
carried out to evaluate the reliability of the experimental condition used for
the in vitro blinatumomab assay. The effector-to-target ratio (1:10),
with exceeding B-precursor cells, was chosen to resemble the invasion of leukemic
blasts in the bone marrow, a condition where a primary resistance to blinatumomab
could result in a dismal outcome for relapsed/refractory CD19 ALL patients
[6, 29, 37, 38, 39]. Besides being administered at relapse, the role of blinatumomab
in at risk patients is now under investigation (e.g., AIEOP-BFM ALL 2017
protocol). In this paper, we have investigated only the 1:10 ratio, but other
effector-to target ratio should be used in further studies for a complete
overview on blinatumomab in vitro effects. Blinatumomab was refreshed on
a daily basis, mimicking more closely drug exposure in patients. Indeed, due to
its short half-life (mean standard deviation: 2.11 1.42 hours in
adults) [40], the drug is administered by continuous infusion in cycles of 4
weeks. The blinatumomab concentration used (1 ng/mL) was in line with steady
state concentration values measured in adult ALL patients during the second cycle
of blinatumomab [41]. The cytofluorimetric assay allowed to appreciate both
blinatumomab-induced effects, i.e., the decreased percentage of B-ALL
immortalized cells combined to the T-lymphocytes increase and activation, at day
+7. However, at this time point, the drug effects were already accomplished since
CD19 cell mortality was almost complete (95%), suggesting that the
ideal timing for investigating simultaneously both drug effects should
fall between days +3 and +7. Interestingly, CD19 cell death was already
observed at day +3 in blinatumomab-treated samples, with a more pronounced effect
in REH than NALM6, contrary to what could be expected considering the reduced
amount of GzB released in the co-culture supernatants and the lack of CD3
activation at this early time point. Several hypotheses could be postulated to
explain such results. It is known that REH cells are more sensitive than NALM6 to
conventional ALL chemotherapy [42], in agreement to what is observed in
ETV6-RUNX1 ALL patients who have good prognostic parameters and a
favorable outcome and in ETV6-PDGFRB patients who failed to reach
remission [43, 44]. In this report, we showed for the first time the different
response of these B-ALL cell lines to the novel immunomodulatory agent
blinatumomab. One hypothesis is that the lower CD19 expressing REH cells
could be more sensitive to cytotoxic stimuli than NALM6 because, in the
balance between cell survival and apoptotic signals, they could be more
susceptible to pro-apoptotic signals induced by the drug. Indeed, cell surface
dynamic analysis revealed that CD19 and BCR come together on plasma membrane
microclusters, and that CD19-defective B-cells show a reduced microcluster
formation and initiation of BCR-dependent signaling, thus affecting the survival
pathway [45]. As a second hypothesis, it could not be ruled out that NALM6 and
REH have a different GzB susceptibility, since GzB-mediated cell death is largely
dependent on a pathway that is regulated by the antiapoptotic protein Bcl-2,
another target gene of BSAP, which is less expressed in BSAP-deficient cells
compared to wild type cells [46, 47]. In our experiments we did not assess
whether NALM6 and REH stimulate T-cell subpopulations (CD8 effector memory
T-cells and CD4 regulatory T cells (Tregs)), differently. CD8 cells
showed a greater and faster blinatumomab-induced cytotoxic capability than other
activated cells, with specific production of cytotoxic factors and cytokines
[48]. A higher percentage of Tregs were associated with unfavorable outcomes in
adult B-ALL patients treated with blinatumomab [49]. This occurred likely because
activated Tregs lead to IL-10 production, resulting in suppression of T-cell
proliferation and in a reduction of the CD8-mediated lysis of ALL cells
[50, 51]. Duell and collaborators demonstrated that CD19 B-ALL blasts and
NALM6 cells induced Treg activation to the same degree [49]. In a recent analysis
at single cell transcriptome resolution, a target cell-dependent mechanism of
T-cell activation by blinatumomab, mediated by the cytokine TNFSF4, was observed
[52]. Whether CD4/ CD8 ratio (and IL-10 or TNFSF4 levels) change
between NALM-6 and REH co-cultures was not established in our experiments.
Finally, upregulation of inhibitory immune checkpoints, mainly PD-L1, has been
recognized as a major mechanism of resistance to BiTE therapy [53], being
increased in relapsed ALL patients and after BiTE treatment in ALLs refractory to
blinatumomab [54]. Future studies could consider PD-L1 surface expression in
NALM6 and REH co-cultures to investigate its contribution to the different
reaction to the drug. All together, these observations suggest that an
in-depth analysis of other activation/inhibitory markers involved in
blinatumomab in vitro susceptibility are needed.
CD19 loss is one of the most significant escape mechanisms of B-ALL blasts
after blinatumomab therapy [55]; however, CD19 negative relapses still represent
a minority of cases, being reported only in 1/3 of patients [56, 57, 58, 59]. Blinatumomab
is given to CD19 B-ALL patients, but there are not studies investigating
the impact of blasts CD19 antigen initial load on treatment success or as
determinant of CD19-positive versus CD19-negative relapses after drug exposure. In
patients treated with tisagenlecleucel (a CD19-specific CAR-T), low tumor burden
is a factor associated with CD19-positive relapses, in contrast to the high
burden associated to CD19-negative ones. It was hypothesized that high amount of
CD19 target (i.e., high tumor burden) could be involved in CAR-T- persistence,
with the drawback of an increased risk of CD19 negative clones selection, able to
escape CAR-T immunosurveillance [60]. For other well-established biological drugs
such as trastuzumab, specifically used for HER2 receptor positive cancers,
therapeutic failure could occur in the presence of high HER2 expression, and
different response rates were observed among subsets of HER2-positive patients
[61]. All together, these evidences suggest that the presence of the molecular
target drives the choice of the biological drug to be used, but a complex and non
linear correlation between the target expression levels and the effective
therapeutic response takes place, particularly for biological therapies that
activate immune cells [5]. Indeed, the therapeutic response is a multifactorial
event that could involve several pathways.
Measurement of the in vitro effects of blinatumomab on primary
mononuclear cells without purification of CD19 or CD3 cells were
performed to assess the assay feasibility on biological material derived from
patients’ bone marrows. Only a single time point measurement was possible due to
the scarce number of alive primary cells recovered after diagnostic procedures
and mononuclear cells isolation; moreover, survival of untreated cells was
compromised over the 7 days of incubation in ~40% of the cases.
Besides these technical limitations, the assay showed a variability in the
blinatumomab in vitro response, and identified co-cultures of patients’
primary cells non-responsive to the BiTE antibody. The blinatumomab in
vitro cytofluorimetric assay could be used to screen the in vivo
patients’ drug response, if its predictive power will be proven; however, this
purpose goes beyond the aim of this paper. Because none of the patients of this
study underwent blinatumomab therapy, a preliminary correlation analysis between
cytofluorimetric in vitro results and in vivo pharmacological
response could not be performed.
5. Conclusions
Results presented in this study are preliminary and far from being exhaustive.
However, the in vitro cytofluorimetric assay here proposed is suitable
for studying the blinatumomab response in CD3:CD19 cell co-cultures,
simulating the condition of lymphoblasts invasion in the bone marrow.
Nonetheless, optimization of the assay is still required, particularly to find
out the ideal time of observation. Screening of a larger panel of B-ALL cell line
models (particularly focusing on those with hyper-activated JAK/STAT pathway
and/or t(17;19) rearrangement) or engineered cells to modulate PAX5
and/or CD19 expression, would help to better understand the
hypothesis-driven question relative to PAX5 or other B-cells genetic
abnormalities contribution in drug response [62, 63]. The in vitro assay
here proposed could represent a model for better understanding the complex
biology of blinatumomab response.
Abbreviations
ALL, Acute lymphoblastic leukemia; BiTE, bispecific T-cell engager; BSAP, B-cell
lineage specific activator protein; GzB, granzyme B; IL-2, recombinant human
interleukin-2; PBMC, peripheral blood mononuclear cells; PBS, phosphate-buffered
saline; Treg, regulatory T cell.
Author contributions
Conceptualization—RF, MG, GS and GD; Data curation—SB, MG and RF; Formal
analysis—SB and RF; Funding acquisition—GD; Investigation—SB, MG, EP;
Writing – original draft—SB and RF; Writing – review & editing—MR, AT, GD
and GS.
Ethics approval and consent to participate
The study was conducted according to the guidelines of the Declaration of
Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of
I.R.C.C.S. Burlo Garofolo (Protocol number CE/V 135; March 5th 2012). Informed
consent was obtained from all subjects involved in the study.
Acknowledgment
The authors gratefully acknowledge the support of Institute for Maternal &
Child Health (I.R.C.C.S) “Burlo Garofolo”, Trieste.
Funding
This work was supported by the Institute for Maternal and Child Health
(I.R.C.C.S) “Burlo Garofolo,” Trieste, Italy (grant number RC 05/2012).
Conflict of interest
The authors declare no conflict of interest.
 Fig. 1.
Fig. 1. Fig. 2.
Fig. 2. Fig. 3.
Fig. 3. Fig. 4.
Fig. 4. Fig. 5.
Fig. 5.