†These authors contributed equally.
Academic Editor: Michael H. Dahan
Background: The aim of this study was to investigate the prevalence and
characteristics of SCCmec genotypes and drug resistance of
methicillin-resistant Staphylococcus aureus (MRSA) isolated from
intensive care units (ICU) at obstetrics & gynaecology departments in a tertiary
hospital. Methods: MRSA obtained from patients admitted to the ICU were
isolated and identified by using the Vitek 2 Compact System with GP21 342 cards.
Antimicrobial susceptibility profiles and MRSA screening were determined by using
the broth microdilution method according to CLSI guidelines. Determination of
resistant genes and SCCmec genotypes were performed by multiplex PCR.
Results: Of the 283 patients evaluated, 120 (42.4%) isolates were
phenotypically and genotypically confirmed to be MRSA. Among 120 strains, 15
(12.5%) strains were SCCmec type II, 96 (80%) strains were
SCCmec type III and 9 (7.5%) strains were undifferentiated type. All MRSA
strains were recognized as multidrug resistant, exhibiting 100% resistance to
cefoxitin and oxacillin, followed by erythromycin and levofloxacin (more than
80% and 90% respectively). Different SCCmec genotypes in MRAS isolates
showed distinct antimicrobial agent patterns. SCCmec type II was highly
resistant to clindamycin (93.3%) with lower resistance to tetracycline (26.7%)
with SCCmec type III being highly resistant to gentamicin
(91.7%). Undifferentiated strains were resistant to Cotrimoxazole (77.8%).
There was a statistical difference among type II, type III and
Undifferentiated strains (P
Methicillin-resistant Staphylococcus
aureus (MRSA) is one of the most common pathogenic bacteria in intensive care
units (ICU) with the majority of isolates demonstrating multidrug resistance
(MDR) which impacts clinical therapy [1, 2]. The resistance mechanism of MRSA is
mainly secondary to the bacteria acquiring a genetic determinant
(methicillin-resistant determinant A and C, abbreviated as mecA or
mecC), which encode penicillin-binding protein PBP2A or
PBP2A
Emerging MRSA and multiple drug resistance are a major public health problem worldwide [5, 6]. They are the most common cause of healthcare-associated infections (HAI) in patients that are admitted to the ICU [7]. HAI occurring in the ICU from MRSA have become particularly problematic since they arise from the treatment received by critically-ill patients [8]. Although evidence suggests a significant increase in the proportion of MRSA hospital infections worldwide, ICU at obstetrics & gynaecology departments have reported only a limited number of MRSA isolates in China [9]. Effective and safe antimicrobial treatment is essential for treating infections in the ICU. Organizational-wide surveillance of infection-derived bacterial isolates and analysis of their susceptibility to different antimicrobial agents provides crucial information for the most effective antimicrobial therapy [10]. Furthermore, a comprehensive analysis of MRSA SCCmec typing of ICU infections and predicting the development trend of drug-resistant strains is critical in evaluating disease prognosis and essential for reducing infection mortality and morbidity in the ICU.
This study was conducted at a university-affiliated hospital in North-East China with approximately 1200 beds. An analysis of retrospective data of MRSA-infected patients in three obstetrics & gynaecology ICUs was conducted during January 2018 to December 2020. A total of 283 obstetric patients were admitted after being transferred from the operating room, general ward and emergency department prior to ICU admission. Patients with concurrent HAI were classified according to infection source such as pneumonia, bloodstream infections, urinary tract infections, surgical site infections or other infections. The pathogenic bacteria associated with a HAI were collected within 48 hours of hospitalization according to the local protocol. Some samples were collected after 48 hours post-hospitalization. The specimens were mainly obtained from sputum or tracheal secretions, pus, blood, ascites, catheters and drainage tubes. Multiple isolates from a single patient were excluded. The isolates from different infected sites of the same patient were also excluded. Approval for collecting clinical samples was granted by the institutional ethics committees of the participating hospital. Informed consent forms were reviewed and signed by all participants before sample collection (Ethical approval number: Protocol Number 2019-01-02).
All isolates were identified as Staphylococcus aureus by conventional
standard procedures and confirmed by VITEK GNI system with GP21 342 cards
(bioMérieuk Vitek Inc., Hazelwood, MO, USA). No repetitive isolates from a
single patient were included. Susceptibility to 14 antimicrobial agents (Bio-Rad,
Hercules, CA, USA) was determined and interpreted by the broth microdilution
method akin to that in the Clinical and Laboratory Standards Institute (CLSI)
criteria. MRSA screening was determined using oxacillin MIC
A series of genes including mecA, femB, mecAa and SCCmec are listed in Table 1. Multiplex PCR amplification and PCR reactions were performed as described elsewhere [12, 13]. PCR products of genes were sent to Sangon Biotech Co., Ltd (Shanghai, China) for sequencing and DNAman software (version 6.0) (Lynnon Biosoft, Vaudreuil, QC, Canada) was used to analyse the sequencing results. Reference strains NCTC 85/2082 was used as standard strains with SCCmec type III and Reference strains NCTC N315 was used as SCCmec type II, respectively.
| Names | Primer sequences (5′ |
Expected length (bp) |
| mecA | GTAGAAATGACTGAACGTCCGATAA | 310 |
| CCAATTCCACATTGATTCGGTCTAA | ||
| femB | TTACAGAGTTAACTGTTACC | 651 |
| ATACAAATCCAGCACGCTCT | ||
| mecAa | GTGAAGATATACCAAGTGATT | 147 |
| ATGCGCTATAGATTGAAAGGAT | ||
| SCCmec I | GCTTTAAAGAGTGTCGTTACAGG | 613 |
| GTTCTCTCATAGTATGACGTCC | ||
| SCCmec II | CGTTGAAGATGATGAAGCG | 398 |
| CGAAATCAATGGTTAATGGACC | ||
| SCCmec III | CCATATTGTGTACGATGCG | 280 |
| CCTTAGTTGTCGTAACAGATCG | ||
| SCCmec IVa | GCCTTATTCGAAGAAACCG | 776 |
| CTACTCTTCTGAAAAGCGTCG | ||
| SCCmec IVb | TCTGGAATTACTTCAGCTGC | 493 |
| AAACAATATTGCTCTCCCTC | ||
| SCCmec IVc | ACAATATTTGTATTATCGGAGAGC | 200 |
| TTGGTATGAGGTATTGCTGG | ||
| SCCmec IVd | CTCAAAATACGGACCCCAATACA | 881 |
| TGCTCCAGTAATTGCTAAAG | ||
| SCCmec V | GAACATTGTTACTTAAATGAGCG | 325 |
| TGAAAGTTGTACCCTTGACACC |
Differences in drug resistance rates of MRSA strains were analysed by Chi-square
test. All drug-resistant data were analysed using SPSS version 13.0 (International Business Machines (IBM) Corp.,
Armonk, NY, USA). Analyses with a value of P
From January 2018 to December 2020, a total of 283 obstetric patients from three
ICU were enrolled in this study to estimate the quantity and types of infections
present in this population. The average age of the patients was 34.88
Both mecA and femB genes were identified by multiplex PCR among all MRSA isolates. The amplicons of mecA and femB genes with a size of 310 bp and 651 bp were illustrated in Fig. 1A, respectively. The findings demonstrated that both mecA and femB genes were found in all MRSA isolates obtained.
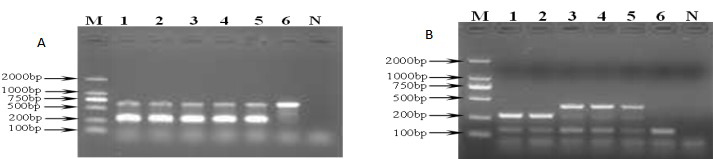 Fig. 1.
Fig. 1.Representative gel showing banding profiles by multiplex PCR analysis in MRSA isolates. (A) Agar gel electrophoresis of mecA and femB detected by multiplex PCR in MRSA isolates. M: DNA molecular weight; 1~5: MRSA isolates from different samples in ICU; 6: MSSA (ATCC25923); N: Negative control. (B) Agar gel electrophoresis of SCCmec types detected by mutiplex PCR in MRSA isolates. M: DNA molecular weight; 1: MRSA isolates SCCmec type III; 2: Reference strains 85/2082 SCCmec type III; 3: Reference strains N315 SCCmec type II; 4–5: MRSA isolates SCCmec type II; 6: MRSA isolates SCCmec undentified type; N: Negative control. MRSA, methicillin resistant Staphylococcus aureus; ICU, intensive care unit.
Out of 120 MRSA isolates,15 isolates belonged to SCCmec type II (12.5%), 96 isolates were SCCmec type III (80%) and 9 strains belonged to an undefined type. PCR patterns are shown in Fig. 1B.
Resistance frequencies of 120 MRSA strains in this study based on CLSI
microdilution demonstrated that they were recognized as multidrug resistant,
exhibiting 100% resistance to cefoxitin and oxacillin, followed by erythromycin
and levofloxacin (more than 80% and 90% respectively). Different
SCCmec genotypes in MRAS isolates showed distinct antimicrobial agent
patterns. SCCmec type II exhibited a high incidence of resistance to
clindamycin (93.3%) but displayed a relatively low prevalence of resistance to
tetracycline (26.7%). SCCmec type III exhibited a high incidence
of resistance to gentamicin (91.7%) while undefined SCCmec types were
resistant to Cotrimoxazole (77.8%). There was a statistical difference among
type II, type III and Undifferentiated strains (P
| Antimicrobial agents | Type II (n = 15) | Type III (n = 96) | Type UD (n = 9) | P value |
| R (%) | R (%) | R (%) | ||
| Cefoxitin | 15 (100.0) | 96 (100.0) | 9 (100.0) | — |
| Oxacillin | 15 (100.0) | 96 (100.0) | 9 (100.0) | — |
| Gentamicin | 0 (0) | 88 (91.7) |
0 (0) |
|
| Clindamycin | 14 (93.3) | 17 (17.7) |
3 (33.3) |
|
| Minocycline | 0 (0) | 0 (0) | 0 (0) | — |
| Teicoplanin | 0 (0) | 0 (0) | 0 (0) | — |
| Linezolid | 0 (0) | 0 (0) | 0 (0) | — |
| Quinupristin | 0 (0) | 0 (0) | 0 (0) | — |
| Compound sulfamethoxazole | 3 (20.0) | 6 (6.25) | 7 (77.8) |
|
| Erythromycin | 14 (92.3) | 87 (90.6) | 8 (88.9) | 1.000 |
| Vancomycin | 0 (0) | 0 (0) | 0 (0) | — |
| Rifampicin | 12 (80.0) | 75 (78.1) | 7 (77.8) | 1.000 |
| Levofloxacin | 14 (93.3) | 94 (97.9) | 9 (100) | 0.4912 |
| Nitrofurantioin | 0 (0) | 0 (0) | 0 (0) | — |
| Note: *P | ||||
Molecular typing of MRSA is an important assay for the epidemiologic investigation and strains of origin in addition to antimicrobial agent selection and therapy. Recent data demonstrate that mecA gene expresses itself in coagulase-negative staphylococcus (CNS) while femB is considered commonly in S. aureus with restricted expression but not present in CNS. Therefore, only when mecA and femB genes are both found to be positive can MRSA be present[14]. This study amplified mecA gene (310 bp) and femB gene (651 bp) from the 120 MRSA strains isolated from clinical samples obtained that were determined to be MRSA.
In the form of a gene complex, mecA gene of MRSA exists in SCCmec that is composed of two gene complexes: mec gene complex and cassette chromosome recombinases (ccr). According to the structures of mec and ccr, SCCmec can be divided to five types [15]. Evidence exists that a majority of early nosocomial infections were generally considered as SCCmec type I while community-acquired MRSA infections were mostly SCCmec type IV and SCCmec type V [16, 17].
In the present study, multiplex PCR amplification was conducted to analyze characteristic genes of SCCmec genotypes. The findings showed that the predominant genotype was SCCmec type III (80%) followed by SCCmec type II (12.5%) while no SCCmec type IV or SCCmec type V was found among MRSA strains in the ICU. As SCCmec type IV and SCCmec type V are mainly community-acquired MRSA infections, it can be verified that community-acquired MRSA infections are not present in our ICU. This finding was in agreement with other reports that 250 clinically isolated MRSA strains were mainly SCCmec type III followed by SCCmec type II in 18 hospitals nationwide in China [18]. An explanation may be the fact that the regional difference provided disparate MRSA with a diversity of SCCmec genotypes, thus leading to distinct drug-resistant patterns. In addition, 9 (7.5%) of 120 isolated MRSA strains of this study were undefined SCCmec strains, probably caused by the selected primers and amplification conditions or that they belong to a novel SCCmec gene, which needs further studies.
In the study, all MRSA strains were recognized as multidrug resistant. They
showed a high incidence of resistance to
Interestingly, a high prevalence of resistance to rifampicin was noted in the
hospital. In fact, rifampicin was seldom administered for the treatment of MRSA.
However, the percentage of rifampicin-resistant MRSA rapidly increased from
15.5% in 2004 to 50.2% in 2008 in China. Several reported that rifampicin
resistance in S. aureus isolates including MRSA was associated with
mutations of rpoB gene (encoding
The treatment of MRSA infection is a very difficult clinical problem. Risk factors for development of an MRSA infection A in the ICU include the following: widespread abuse, misuse, or overuse of antibiotics; the frequent renewal of antibiotics; increasing quantity and variety of pathogens present in the ICU; the diagnosis and progression of critical illnesses; postsurgical recovery; the use of invasive medical devices; and prolonged stay in the ICU [22, 23]. Patients should be monitored for drug resistance within S. aureus so as to provide a basis for determining appropriate therapy. Meanwhile, hospitals are expected to allocate dedicated ventilators, oxygen, transfusion system, sphygmomanometer and thermometer to each patient and to disinfest the equipment after being used by each person. It is necessary to train the medical staff to infection prevention since researches verify that the hands of the medical staff are an important medium to spread MRSA, which means that hand washing is a vital way to block the spread of MRSA [22, 23]. Also, attention should be paid to ventilation and air purification. Moreover, antibiotics should be used carefully in clinical applications to prevent drug-resistant strains along with disinfection and proper isolation when indicated in order to reduce cross infection of S. aureus [24].
This study has a few limitations. First, genotypic or molecular data including Panton-Valentine leukocidin genes among all unidentified strains were not determined. Second, the antibiotic sensitivity of MRSA isolates to Daptomycin was not included. Future research may consider focusing on genetic types and the mechanisms for transmission.
The findings of this study indicate that the MRSA isolates are circulating in the ICU at obstetrics & gynaecology departments and constitute a major source of infection at a large hospital in China. This study also found that vancomycin may be a reasonable choice in the treatment of MRSA isolates. There is a strong need for increased hospital-wide surveillance and the development of adequate infection prevention strategies.
ICU, intensive care unit; CLSI, the Clinical and Laboratory Standards Institute; HAIs, healthcare-associated infections; MRSA, methicillin-resistant Staphylococcus aureus.
ZHH conceived, designed the experiments and wrote a draft manuscript. BXL and MCL analyzed, interpreted the results of the experiments and revised the manuscript. PYL performed the experiments. XM collected the clinical data. All authors read and approved the final manuscript.
Ethical approval for collecting clinical samples was received by the institutional ethics committees of the participating hospital. Informed consent forms were reviewed and signed by all participants before samples collection (Ethical approval number: Protocol Number 2019-01-02).
We thank three anonymous reviewers for excellent criticism of the article.
The present project was a part of National Natural Science Foundation of China project (81402979), the Jilin Science and Technology Development Program (20190304101YY and 20190301014NY); the Health and Family Planning Commission of Jilin Province (2018J098) also supported the project.
The authors declare no conflict of interest.
