1 Department of Botany, Government College University, 5400 Lahore, Pakistan
2 Department of Zoology, Government College University, 5400 Lahore, Pakistan
3 Department of Botany, Lahore College for Women University, 5400 Lahore, Pakistan
4 Faculty of Pharmacy, College of Pharmacy, University of the Punjab, 5400 Lahore, Pakistan
5 MARE - Marine and Environmental Sciences Centre, Department of Life Sciences, University of Coimbra, Calçada Martim de Freitas, 3000-456 Coimbra, Portugal
6 College of Forestry, Hainan University, 570228 Haikou, Hainan, China
Academic Editor: Baohong Zhang
Abstract
Aqueous extracts of two red seaweeds Halymenia porphyriformis and
Solieria robusta were used to synthesize green silver nanoparticles.
These biogenic nanoparticles were tested against four strains of oral pathogenic
bacteria which cause tooth decay or cavities. Staphylococcus aureus
(MT416445), Streptococcus viridans (MT416448), Lactobacillus
acidophilus (MT416447) and Lactobacillus brevis (MT416446) were used.
Characterization of AgNPs was done by UV-Visible spectroscopy, SEM, XRD and FTIR.
XRD analysis revealed the crystalline nature of the particles. The size analysis
by XRD of the green AgNPs by H. porphyriformis indicated it had smaller
particles, 15.23 nm, when compared to AgNPs by S. robusta (17 nm). Both
green synthesized silver nanoparticles showed moderate antibacterial activity
against all strains of bacteria, except L. acidophilus. Both particles
showed their maximum zone of inhibition against L. acidophilus at a
lower concentration of 50 and 100
Keywords
- red algae
- silver nanoparticles
- oral pathogenic bacteria
- tooth decay
The synthesis of nanoparticles using physical and chemical methods has been well exhibited in various studies, but these methods are less ecofriendly, and more expensive and poisonous [1, 2, 3]. So, there is a need to adapt a natural route, for example plant extract, bacteria [4], fungi [5], enzymes [6], and algae [7], for the production of ecofriendly metal nanoparticles (NPs). All these routes are presently under an extensive research process [8]. Due to the conducive behavior towards biological processes, they have widely been used in the medical field, particularly against bacterial activity [9].
The extensive utilization of antibiotics to control bacterial infections is causing microbes to become resistant against multiple drugs. Now the leading problem which is faced around the world is to provide a sound, effective and natural treatment against microbial strains which have become resistant to drugs. Hence, the application of NPs in the field of medicine is considered to be an alternative bactericide, which showed remarkable bactericidal activity. As these NPs disrupt the cell membrane of bacteria and generate reactive oxygen species (ROS) in order to destroy the living machinery of the cell, they have a wide range of antibacterial activity against both gram-negative and gram-positive bacteria [10, 11]. The antibacterial activity of various NPs synthesized from algae has been evaluated against bacterial pathogenic strains. It has been investigated that the bacterial strain of Raoultella planticola (formerly Klebsiella planticola), Pseudomonas aeruginosa, Bacillus subtilis, and other Bacillus sp. [11, 12, 13] showed decrease in growth against AgNPs synthesized from brown seaweed Padina tetrastromatica. In another investigation, AgNPs of colloidal shaped particles prepared from marine green algae Caulerpa serrulata showed remarkable bactericidal activity at very low concentration against Shigella sp., Staphylococcus aureus, Escherichia coli, P. aeruginosa and Salmonella typhi [14, 15, 16].
In the present investigation, AgNPs synthesized from red algae Halymenia porphyriformis and Solieria robusta (Rhodophyta) [17] were examined against dental pathogens. The phytochemical screening of various species of this genus confirms the presence of phytochemical constituents including saponins, flavonoids, terpenoids, carbohydrates, glycosides, protein, fats and alkaloids [18, 19]. It has been reported that alkaloids are abundantly found in Rhodophyta and the presence of these alkaloids plays an important role in the pharmaceutical field as an antibacterial [20, 21, 22], antifungal and insecticidal agent, along with its role in neurotransmission, neuromodulation, growth regulation, antioxidant and cytotoxicity [23, 24]. Flavonoids are known to be a nature’s delicate drug, all these phytochemicals enhancing the antimicrobial and antioxidant activities [25, 26, 27].
Along with this, silver is known to be a toxic agent for microbes, as Ag ions cause structural and morphological changes in bacterial cells and thus disturb the biochemical machinery of the cell, which, in turn, causes cell death [28]. Out of all known metal nanoparticles, AgNPs have the highest bactericidal effects [29, 30] due to the formation of free radicals and subsequent free radical which induces membrane disruption. The antibacterial activity of AgNPs has been studied widely with human pathogenic agents [31, 32, 33]. There are innumerous microbial species in nature which are acidogenic and are involved in the origination and continuation of dental diseases in humans [34, 35, 36]. Streptococcus is considered to be one of the most common disease-causing agents in oral diseases [37, 38], especially plaque, because of its anomalous properties of fermenting sugars and synthesizing extracellular polysaccharides [39, 40]. The major disease-causing agents of root caries and coronal caries are S. sobrinus and S. mutans, and some acid tolerant species including S. sanguis, S. gordonii and S. oralis. The second most common etiologic agents are the species of Lactobacillus [41]. So in the present study it was studied to check the effect of different silver nanoparticles against bacteria inhabiting dental environment.
Samples were collected from the sea coast of Karachi. Two specimens of red algae, i.e., Halymenia porphyraeformis and Soleria robusta were collected using scoop and then placed them in jars separately and labelled them accordingly. Collected samples were brought to the laboratory for further study.
Collected samples of H. porphyraeformis and S. robusta were washed properly with tap water so that excessive material washed away. After washing samples air dried by spreading them under the shade. After drying each sample was separately grinded into fine powdered form by using electrical grinder. After grinding samples were stored in the air tight containers with their respective label.
One gram of Halymenia porphyriformis was taken and grounded into fine
powder. The mixture was heated on a hot plate at the temperature of
60–65 ºC for 10–15 min in 100 mL of distilled water.
Filtration of the mixture was then done by using Whatman n
 Fig. 1.
Fig. 1.Aqueous extract of H. porphyriformis (A–C) and of S. robusta (D–F).
Two flasks were taken, one contained 20 mL extract of S. robusta and the other contained 20 mL extract of H. porphyriformis. For the preparation of the reaction mixture, the extract and 0.01 M silver nitrate were dissolved in the ratio of 8:1. The formation of AgNPs was observed by the appropriate color change for 24 h (Fig. 2). The reaction mixture was centrifuged at 12,000 rpm for 15 min. To obtain silver nanoparticles in powder form, the supernatant was discarded and the pellet containing silver nanoparticles settled down. It was collected in the separate centrifugation tube. The pellet obtained as a result of centrifugation was air dried by placing the centrifugation tubes open for about 3–4 weeks [42, 43].
 Fig. 2.
Fig. 2.Reaction mixture (A) of 20% H. porphyriformis at initial stage (B–D). Reaction mixture of 20% H. porphyriformis after 4, 16 and 24 h. Reaction mixture (E) of 20% S. robusta at initial stage (F–H). Reaction mixture of 20% S. robusta after 4, 16 and 24 h.
Absorption of reaction mixture of silver nitrate and green extract was taken from 320 nm to 700 nm using UV-Vis spectrophotometer in order to confirm the synthesis of NPs. Scanning Electron Microscopy (SEM) JEOL JSM-6480LV, X-ray diffraction analysis (XRD), GLP, ISO17025, and Fourier Transmission Infrared (FTIR) IR Prestige-21, SHIMADZU, were used to characterize the composition, shape, surface topography of the substance and functional groups attached with AgNPs [42].
To evaluate the antibacterial activity of synthesized AgNPs, the following strains of bacteria were checked by using MIC and MCB methods. Isolated strains of dental pathogens were Staphylococcus aureus (MT416445), S. viridans (MT416448), Lactobacillus acidophilus (MT416447) and L. brevis (MT416446). Cultures of selected bacterial strains were prepared in Petri plates [44]. For culturing of bacterial strains, LB medium was used.
The suspensions from pure cultures of the selected bacteria were prepared by
growing all pathogenic strains in a single colony with sub culturing in tryptone
broth to reach 105–106 CFU. 10 mg of dried AgNPs were added to 10 mL of
distilled water and then the mixture was sonicated in order to obtain a
homogenous suspension of AgNPs at concentration of 1000
To evaluate the antibacterial activity of AgNPs of S. robusta and
H. porphyriformis, the serial dilution method was used. For this
purpose, a stock solution of 10 mg/mL of NPs was used. In order to prepare the
serial dilution of AgNPs with water as solvent, 0.4 mL from stock solution was
taken and then added in 19.6 mL of distilled water to make the first dilution of
200
For the evaluation of antibacterial activity of synthesized silver
nanoparticles, a well was formed in the center of inoculating Petri plates by
using a cork borer of 3 mm in diameter. Three different concentrations (200, 100
and 50
After 24 h, a zone of inhibition was observed in all the incubating Petri
plates. Readings were taken in millimeter, or the determination of antibacterial
activity of AgNPs of H. porphyriformis and S. robusta serial
dilution method was used [46]. For the evaluation of MIC and MBC, three different
concentrations of AgNPs were used, i.e., 200, 100 and 50
Statistical analysis of the obtained data was done with SPSS version 22.0
statistical package for MS Window (SPSS Inc., Chicago, IL, USA). Mean values and
standard deviation of ZOI of intra group was analyzed using paired ‘t’
test while intergroup mean and standard deviations were analyzed using One way
ANOVA. Statistical significance was analyzed using Tuck’s Post Hoc HSD test
(p
After the formation of the reaction mixture, a change in color was observed in both experimental groups. The reaction took time for completion of about 24 h. The color of the extract started changing from colorless to reddish brown after 24 h. The reddish-brown color specified the formation of silver nanoparticles.
Similar procedure was followed for the preparation of silver nanoparticles of S. robusta and H. porphyriformis.
UV-Vis spectrometer analysis:
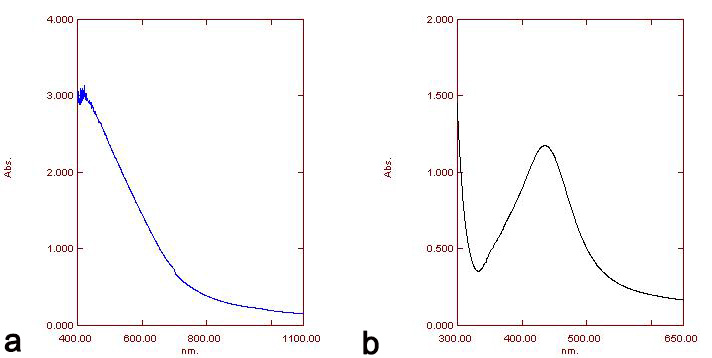
The pellet was used for characterization via UV-Visible spectrophotometry in the wavelength range of 300–700 nm. UV spectrophotometry of AgNPs of both specimens showed maximum absorption at 420 nm (Fig. 3).
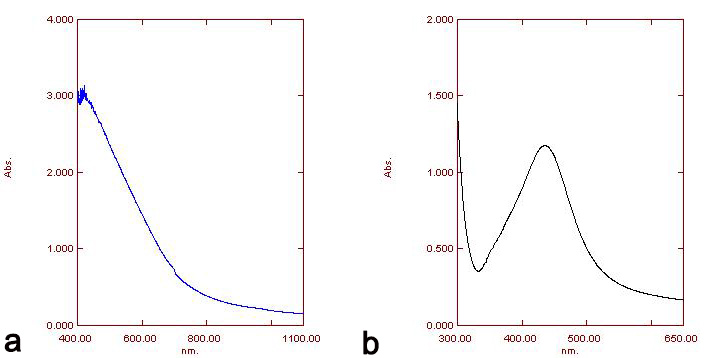 Fig. 3.
Fig. 3.Absorption spectra UV-vis of AgNPs of H. porphyraeformis (a), and of AgNPs of S. robusta (b).
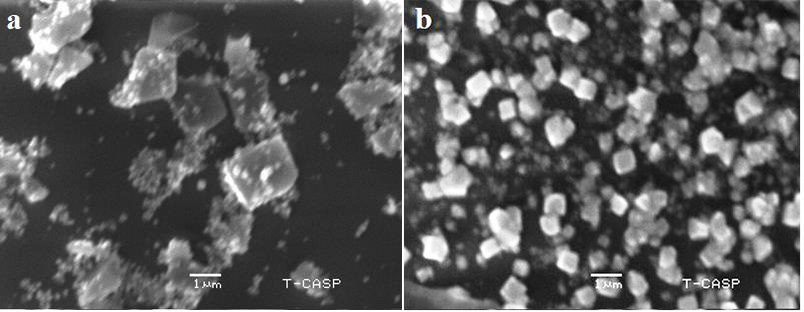
The morphology and structure of nanoparticles were observed by SEM analysis. The AgNPs of H. porphyriformis were cubic and spherical in shape while the particles synthesized from S. robusta were cubic and cylindrical in shape (Fig. 4). It has been proven that improved penetration due to high magnfication enables to observe the better surface morphology of nanoparticals which is only possible in SEM [47, 48, 49, 50, 51, 52].
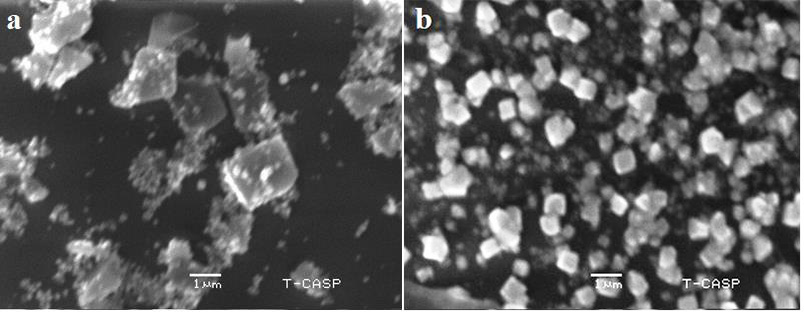 Fig. 4.
Fig. 4.SEM images of AgNPs of H. porphyriformis (a), and of AgNPs of S. robusta (b).
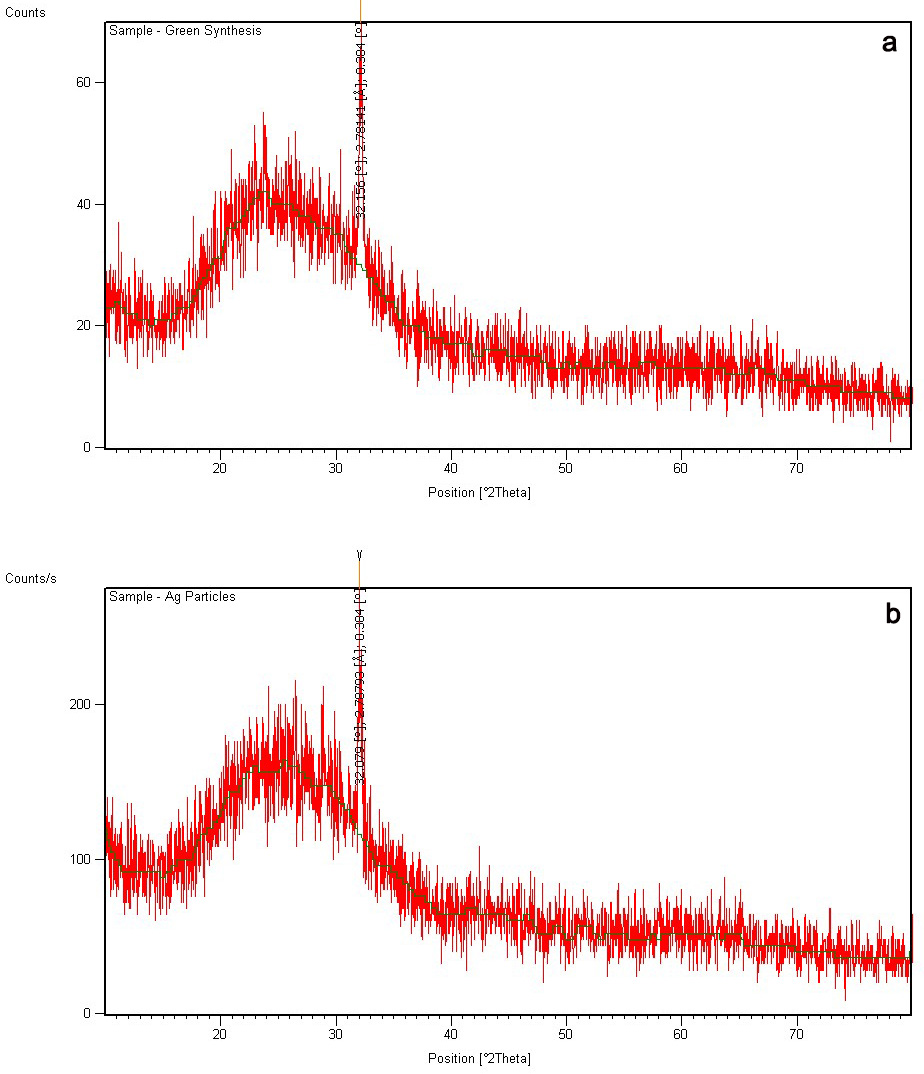
The XRD analysis of both green synthesized nanoparticles was done. The maximum
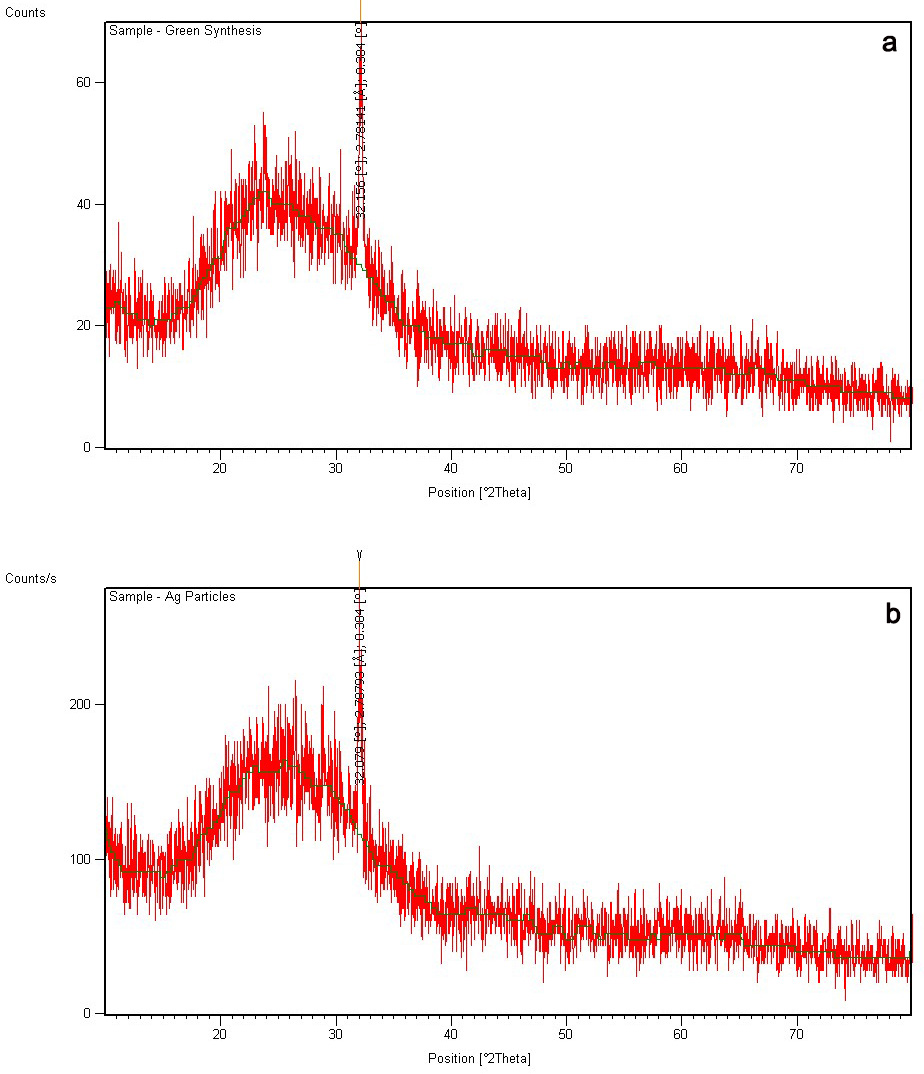 Fig. 5.
Fig. 5.XRD pattern of AgNPs of H. porphyriformis (a), and of AgNPs of S. robusta (b).
The size of the green AgNPs of H. porphyriformis was calculated as 15.23 nm while AgNPs of S. robusta was 17 nm [42].
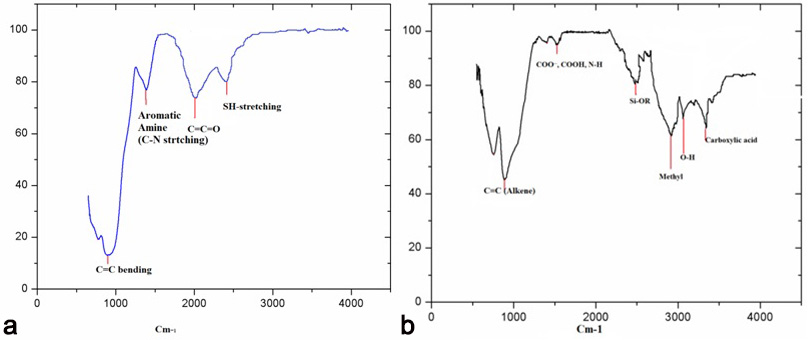
The FTIR analysis indicated that C=C bending (Fig. 6), S-H groups and aromatic functional groups were attached with both green synthesized AgNPs.
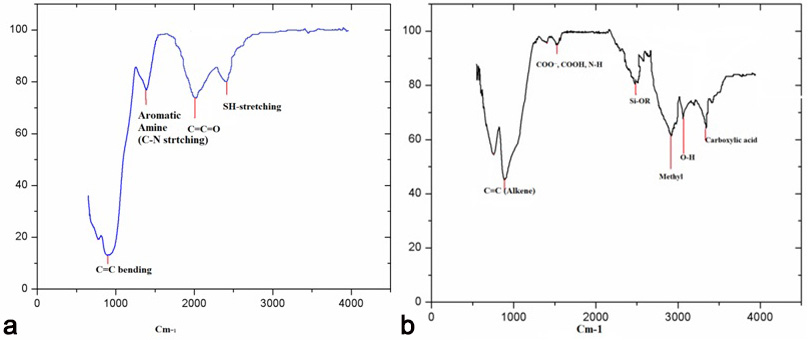 Fig. 6.
Fig. 6.FTIR spectra of AgNPs of H. porphyriformis (a), and of AgNPs of S. robusta (b).
Staphylococcus aureus (MT416445), Streptococcus viridans (MT416448), Lactobacillus acidophilus (MT416447) and Lactobacillus brevis (MT416446). Staphylococcus aureus (MT416445), Streptococcus viridans (MT416448), Lactobacillus acidophilus (MT416447) and Lactobacillus brevis (MT416446).
In this study, MIC and MBC of AgNPs of S. robusta and H.
porphyriformis were investigated against four bacterial strains of dental
pathogens. Agar media cultures were used to investigate antibacterial activity
[53]. Results were statistically significant at a value p
To measure the MBC three different concentrations of 200, 100 and 50
| Pathogenic bacteria | Zone of inhibition in mm (Mean | ||||
| Positive control (Amoxillin) | Negative control | Different concentrations of green synthesized AgNPs of S. robusta | |||
| 200 µg | 100 µg | 50 µg | |||
| Staphylococcus aureus (MT416445) | 4.6 |
0 | 5 |
4.3 |
3.3 |
| Streptococcus viridans (MT416448) | 4.7 |
9.0 |
4.3 |
2.3 |
4 |
| Lactobacillus acidophilus (MT416447) | 8.9 |
6.6 |
2.1 |
9.6 |
10.6 |
| Lactobacillus brevis (MT416446) | 9.7 |
0 | 6.9 |
7.6 |
7.3 |
| Pathogenic bacteria | 200 µg/mL | 100 µg/mL | 50 µg/mL |
| Staphylococcus aureus (MT416445) |  |
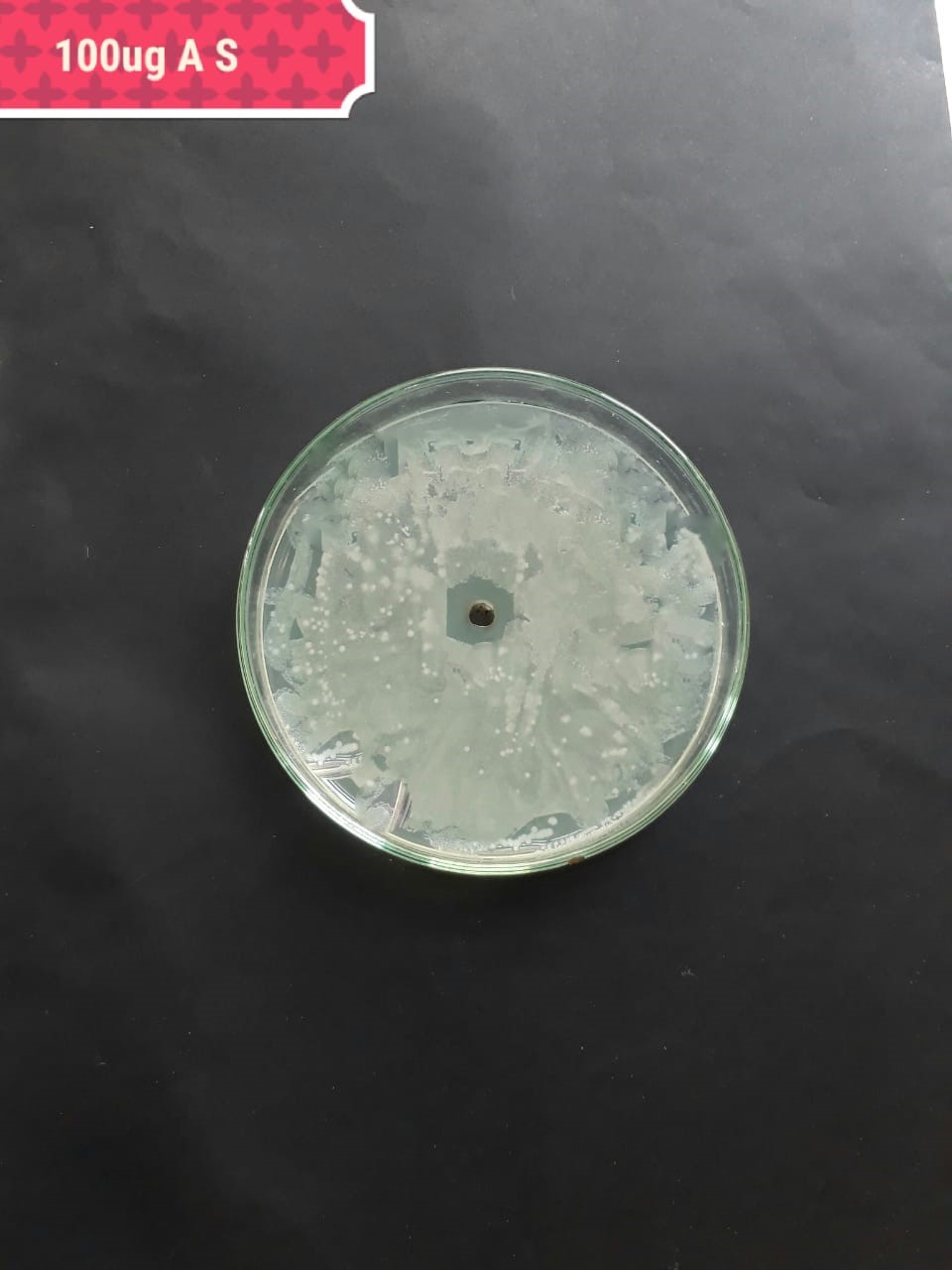 |
 |
| (MT416448) | 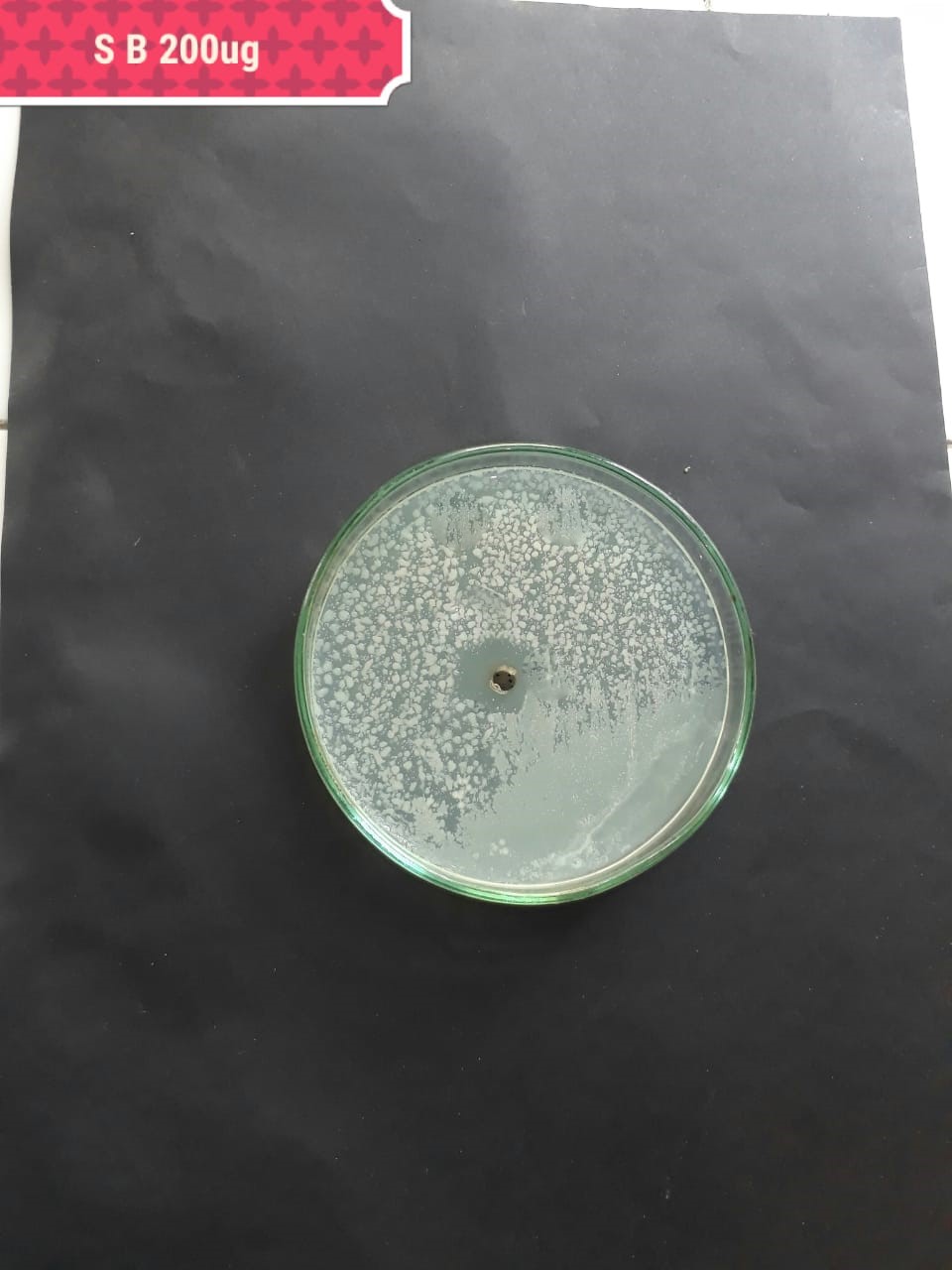 |
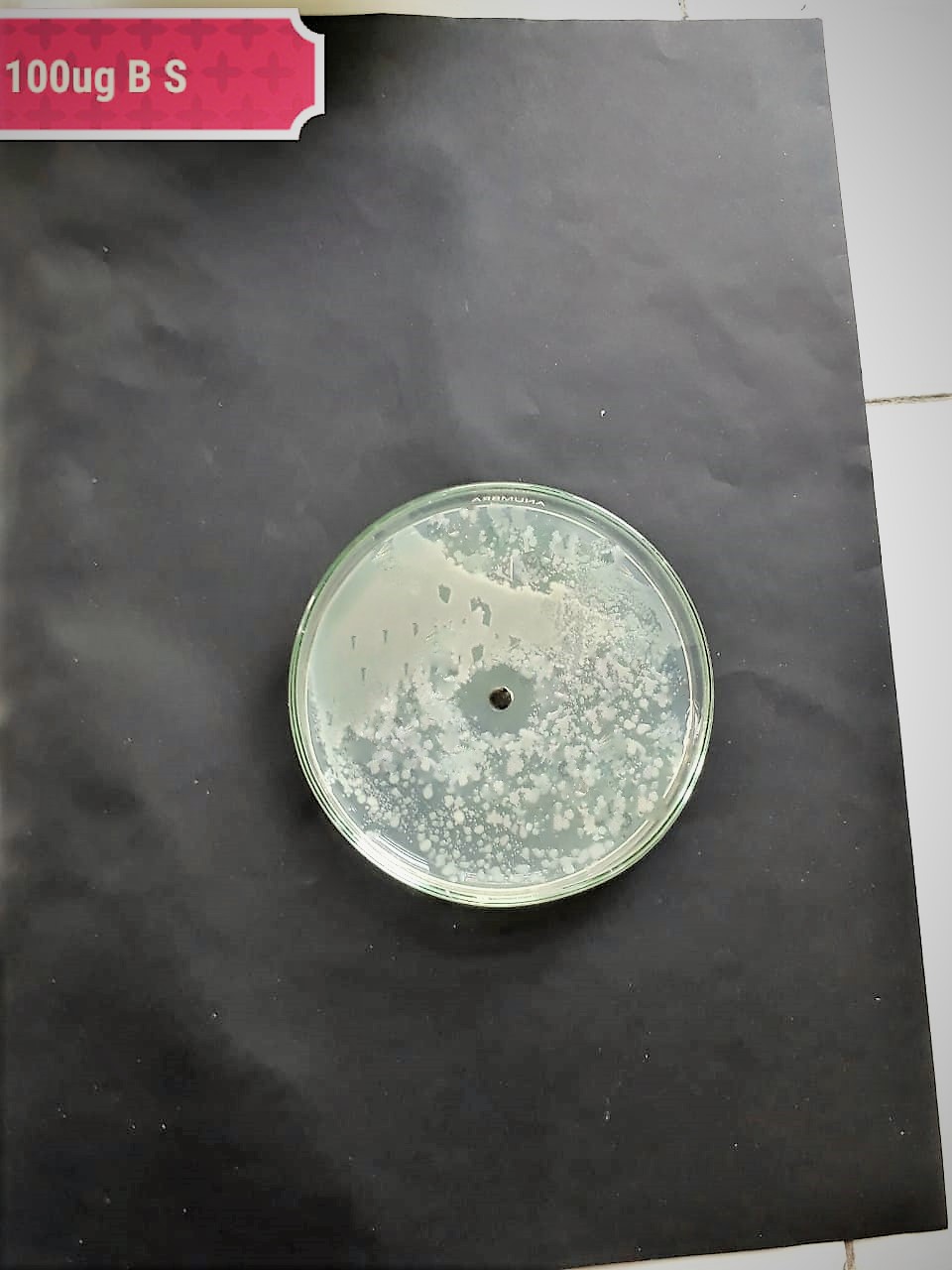 |
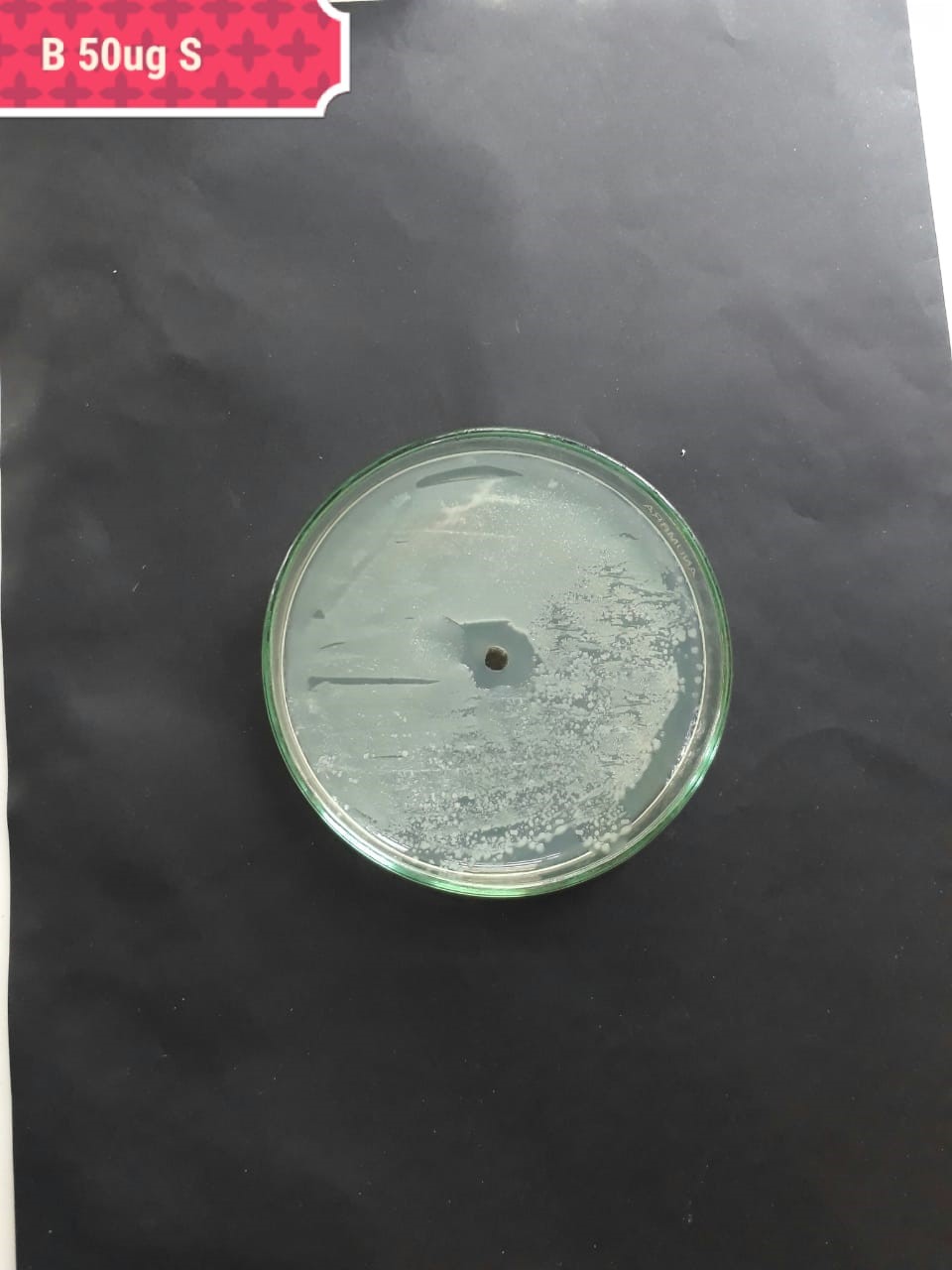 |
| Lactobacillus acidophilus (MT416447) |  |
 |
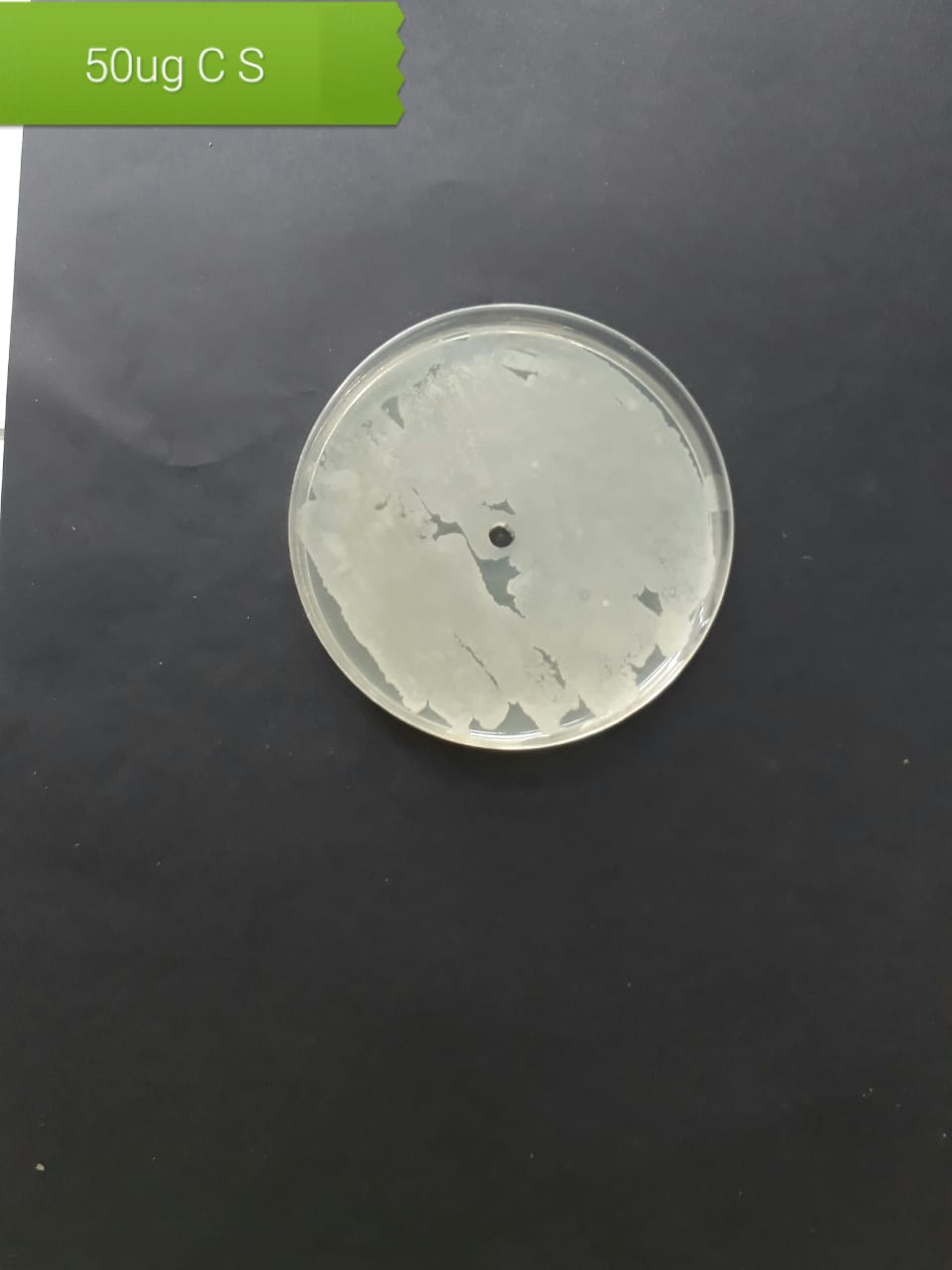 |
| Lactobacillus brevis (MT416446) |  |
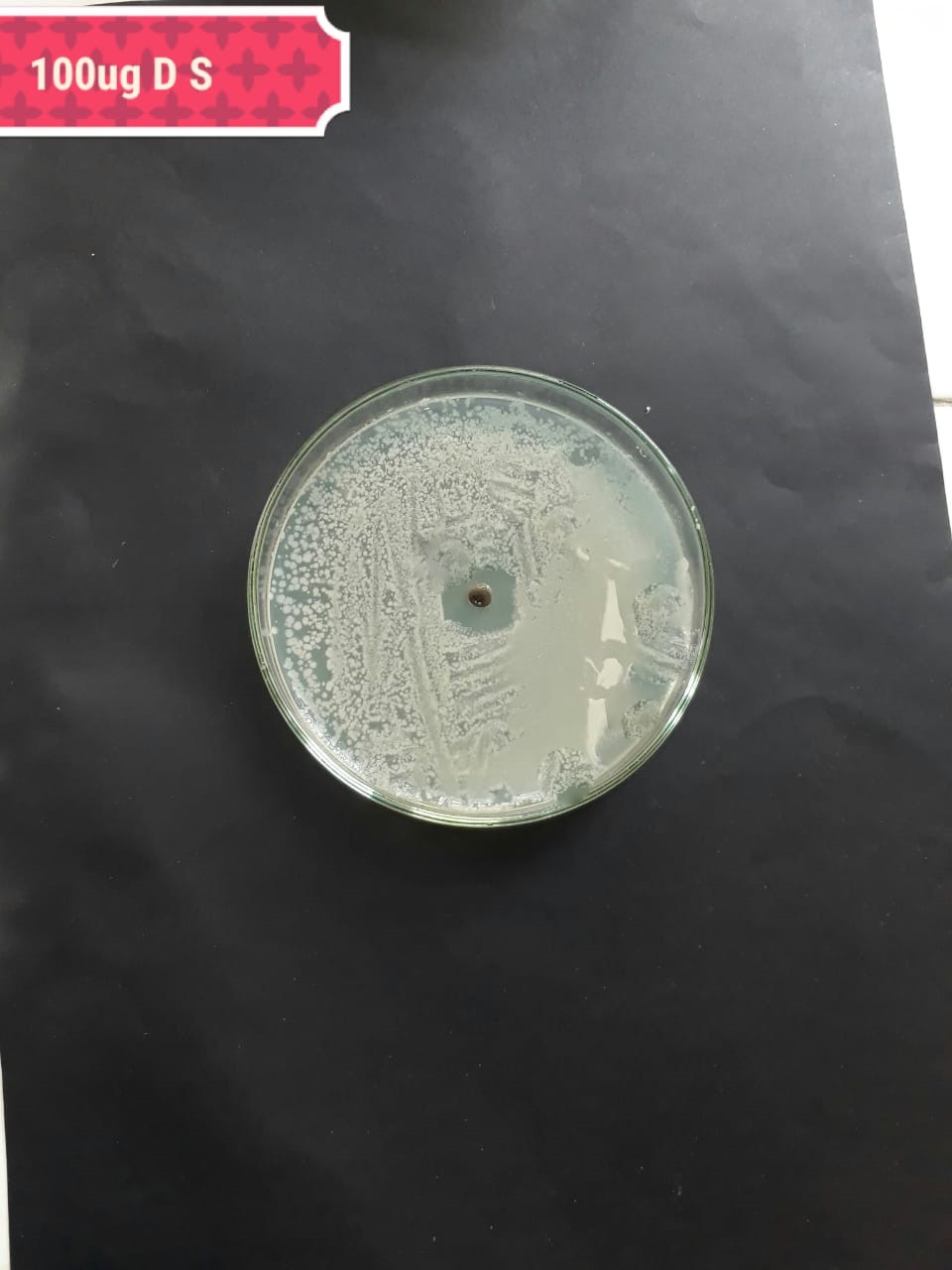 |
 |
In the case of AgNPs synthesized from H. porphyriformis, maximum ZOI at
concentration of 200
| Pathogenic bacteria | Zone of inhibition in mm (Mean | ||||
| Positive control (Amoxillin) | Negative control | Different concentrations of green synthesized AgNPs of H. porphyraeformis | |||
| 200 µg | 100 µg | 50 µg | |||
| Staphylococcus aureus (MT416445) | 4.6 |
0 | 4.6 |
5 |
3.33 |
| Streptococcus viridans (MT416448) | 4.7 |
9.0 |
8.9 |
5.3 |
7.0 |
| Lactobacillus acidophilus (MT416447) | 8.9 |
6.6 |
11.6 |
12.5 |
11.9 |
| Lactobacillus brevis (MT416446) | 9.7 |
0 | 10 |
11 |
11.6 |
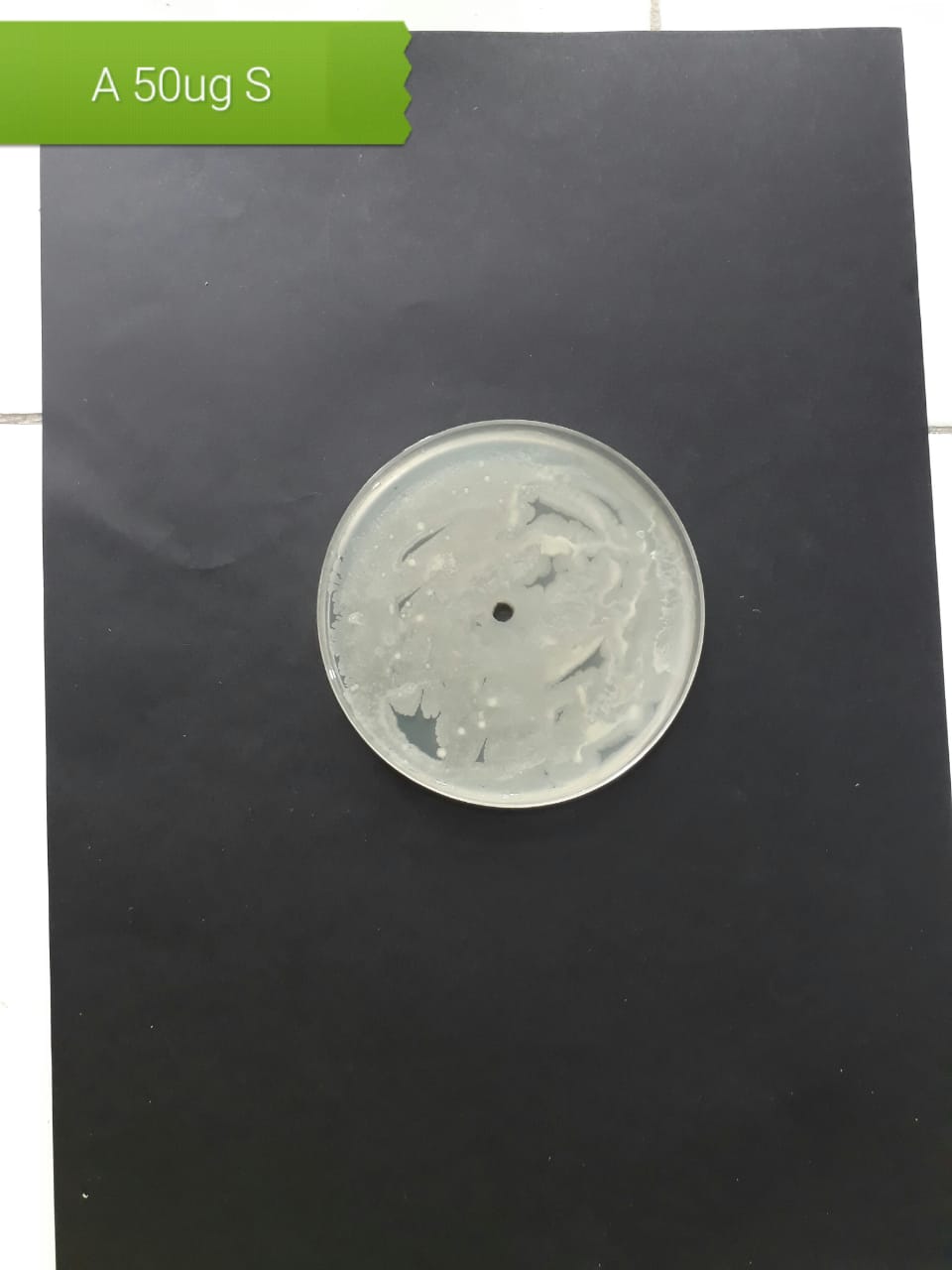
| Pathogenic bacteria | 200 µg/mL | 100 µg/mL | 50 µg/mL |
| Staphylococcus aureus (MT416445) | 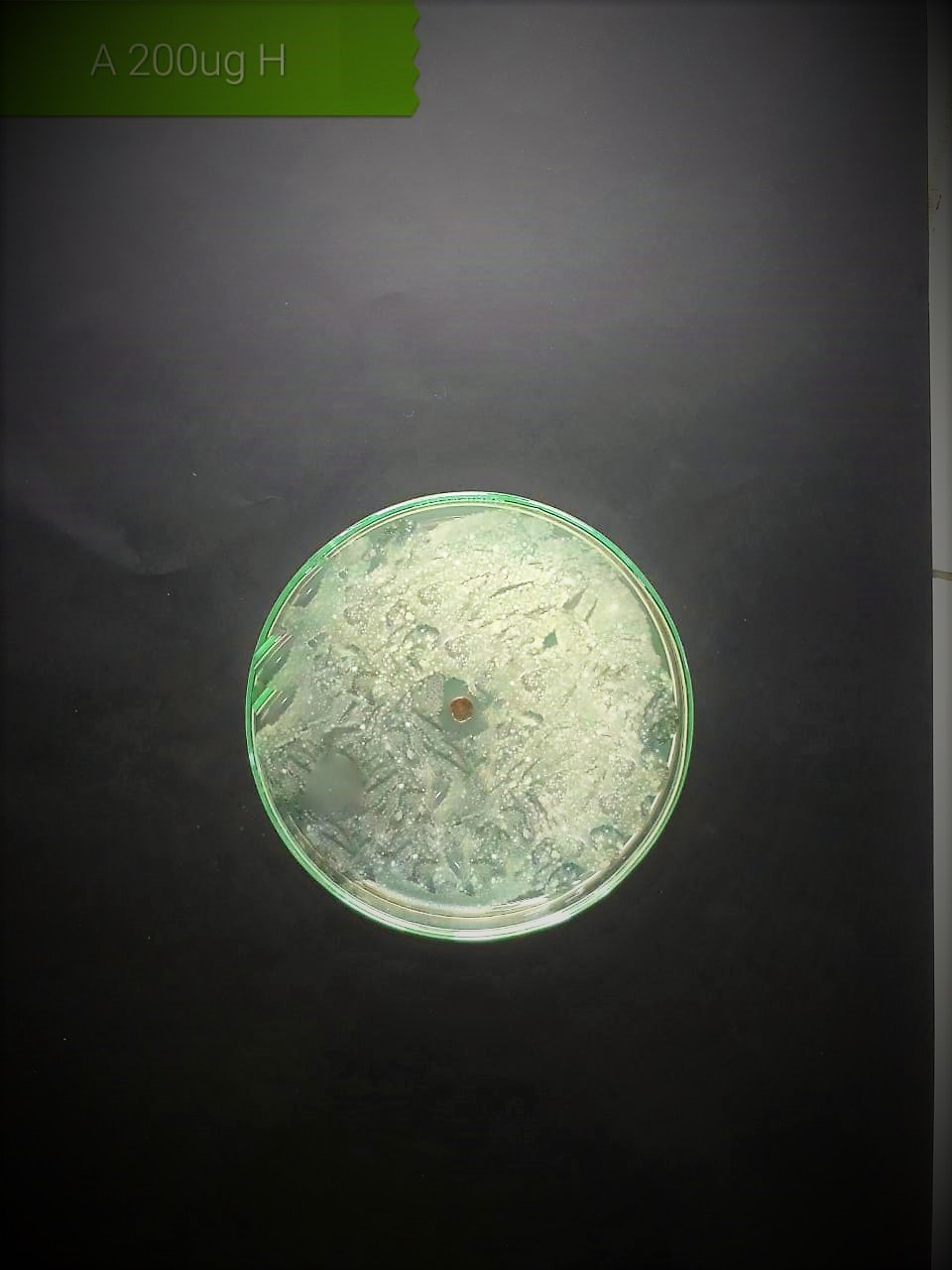 |
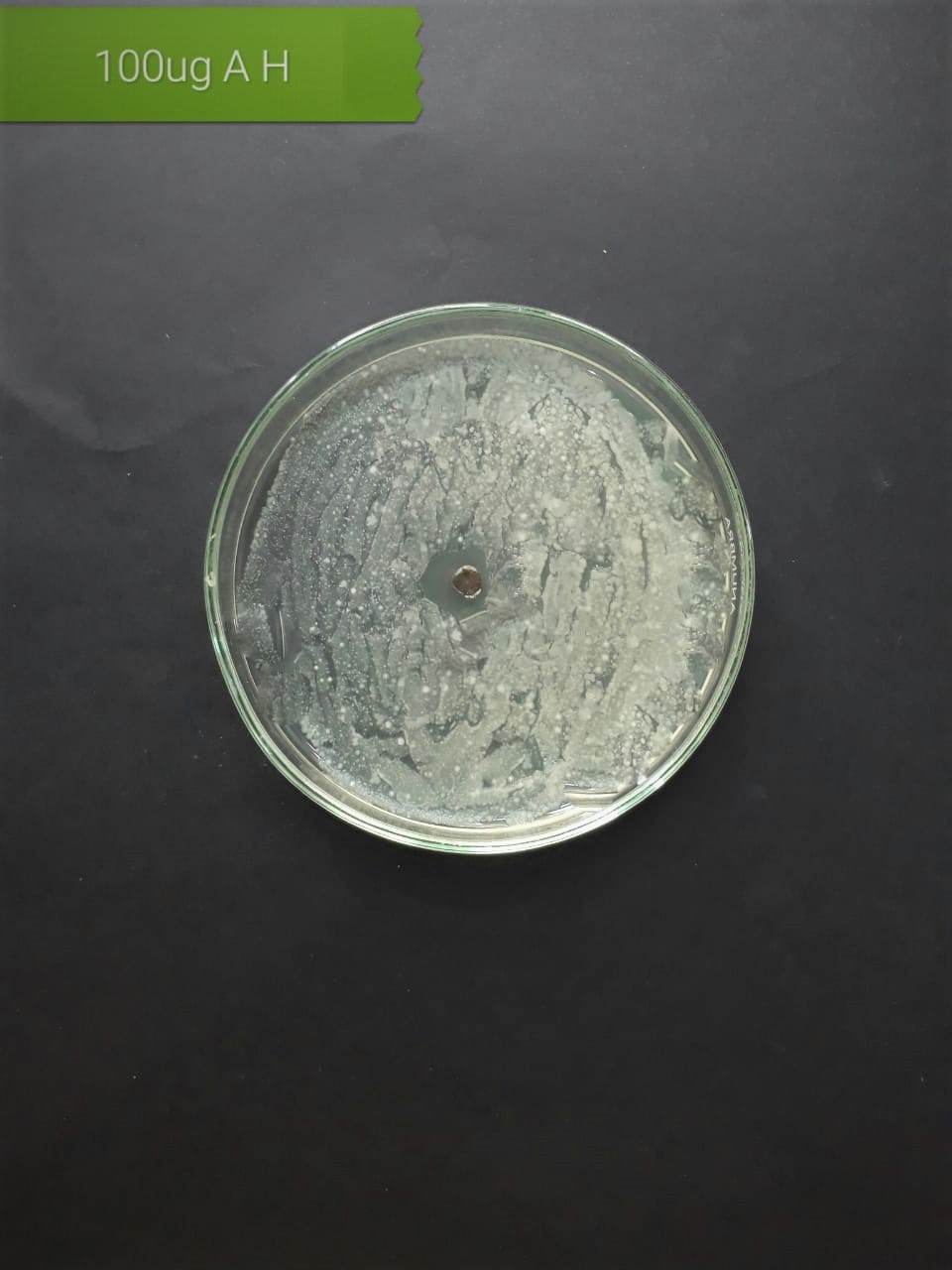 |
 |
| Streptococcus viridans (MT416448) | 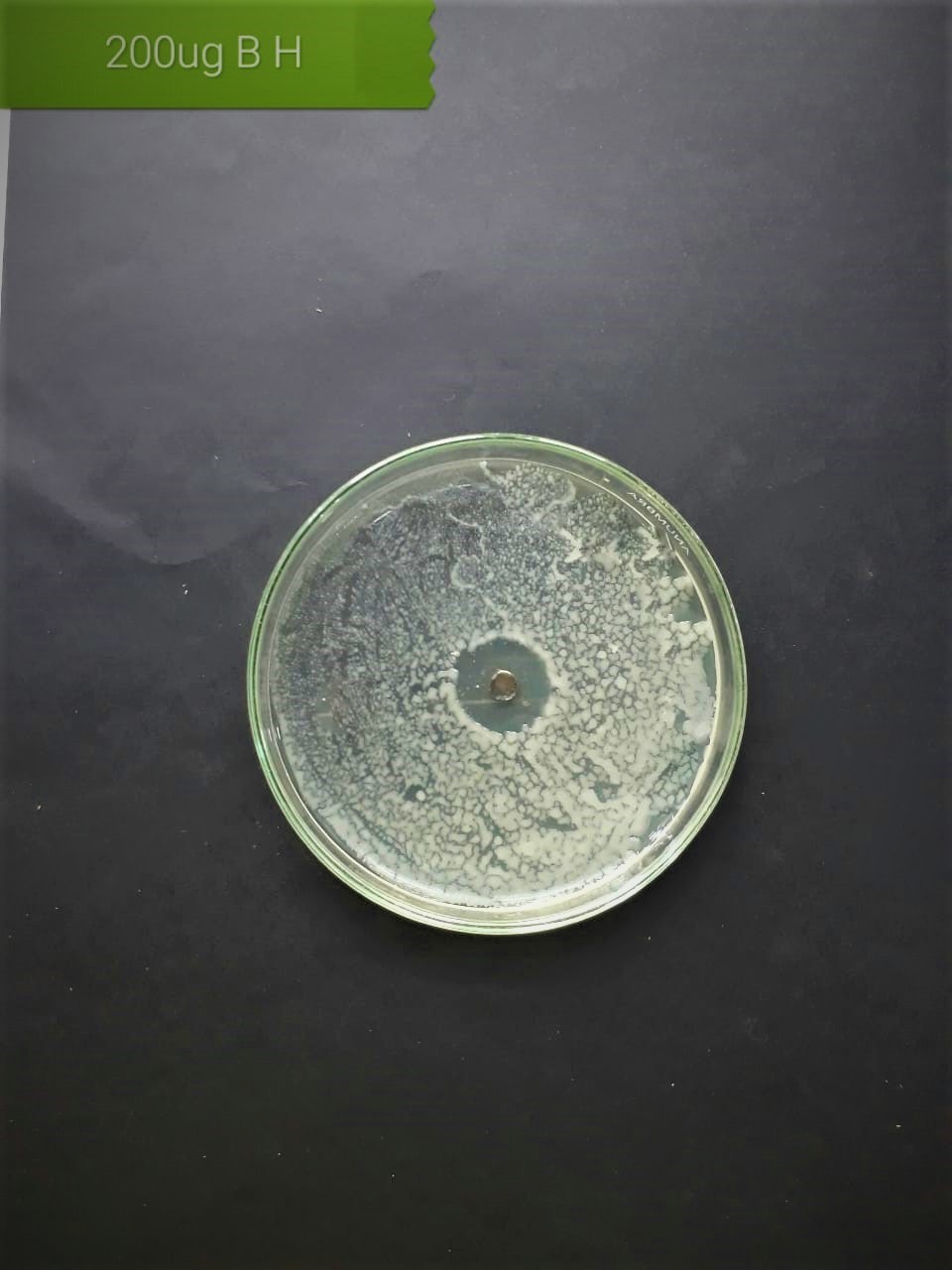 |
 |
 |
| Lactobacillus acidophilus (MT416447) | 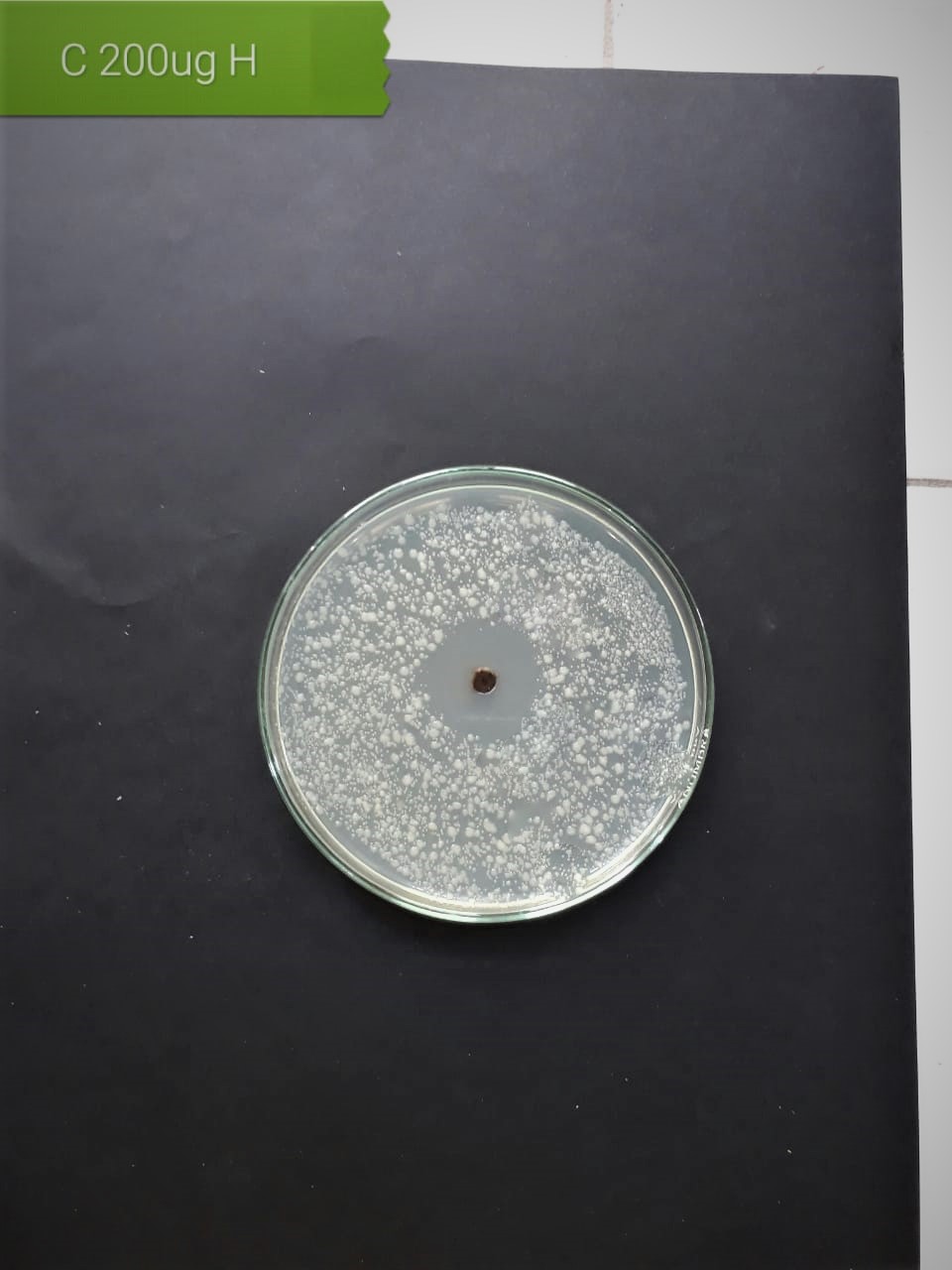 |
 |
 |
| (MT416446) | 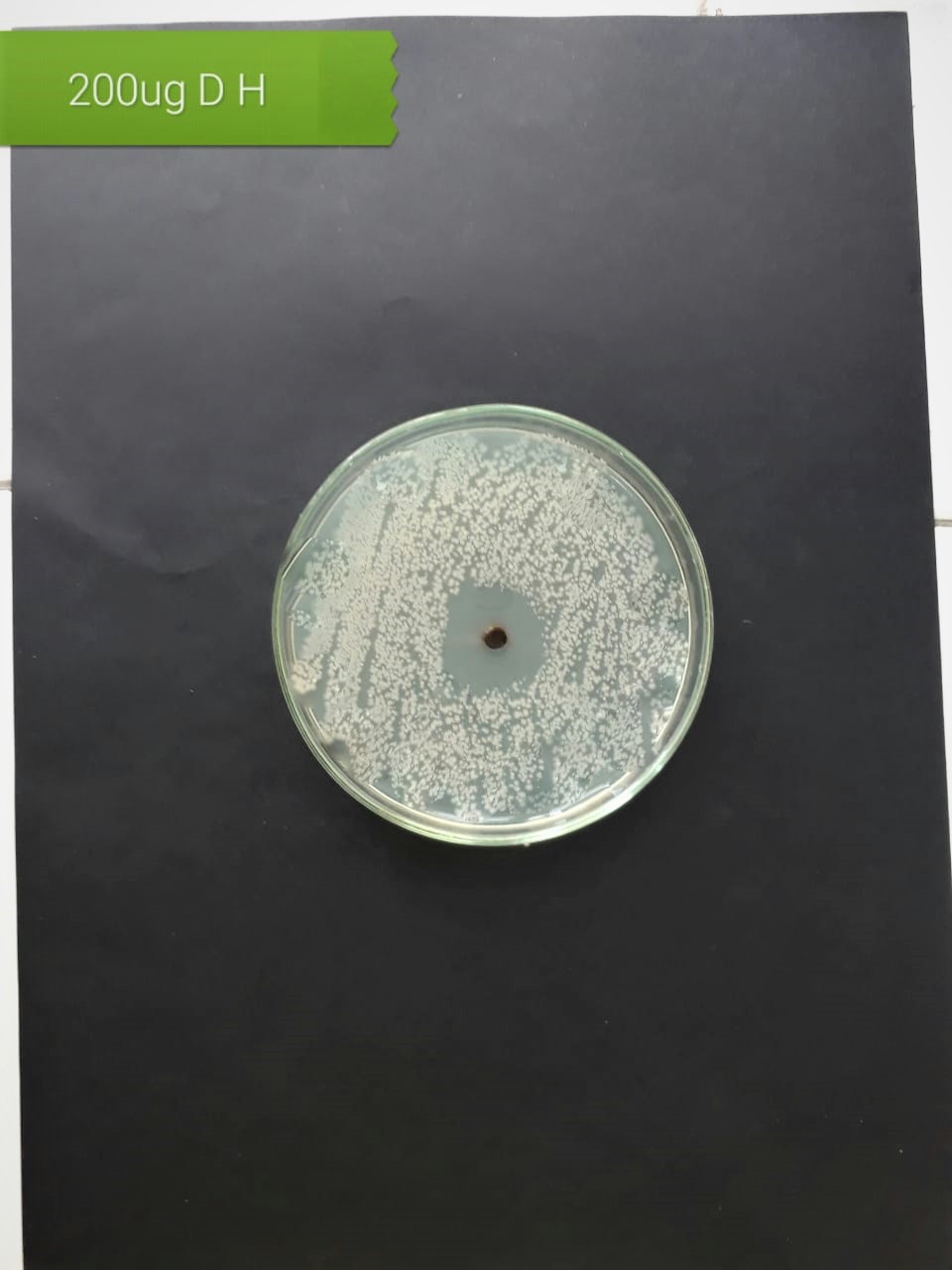 |
 |
 |
| Bacterial strains | Positive control | Negative control |
| Staphylococcus aureus (MT416445) | 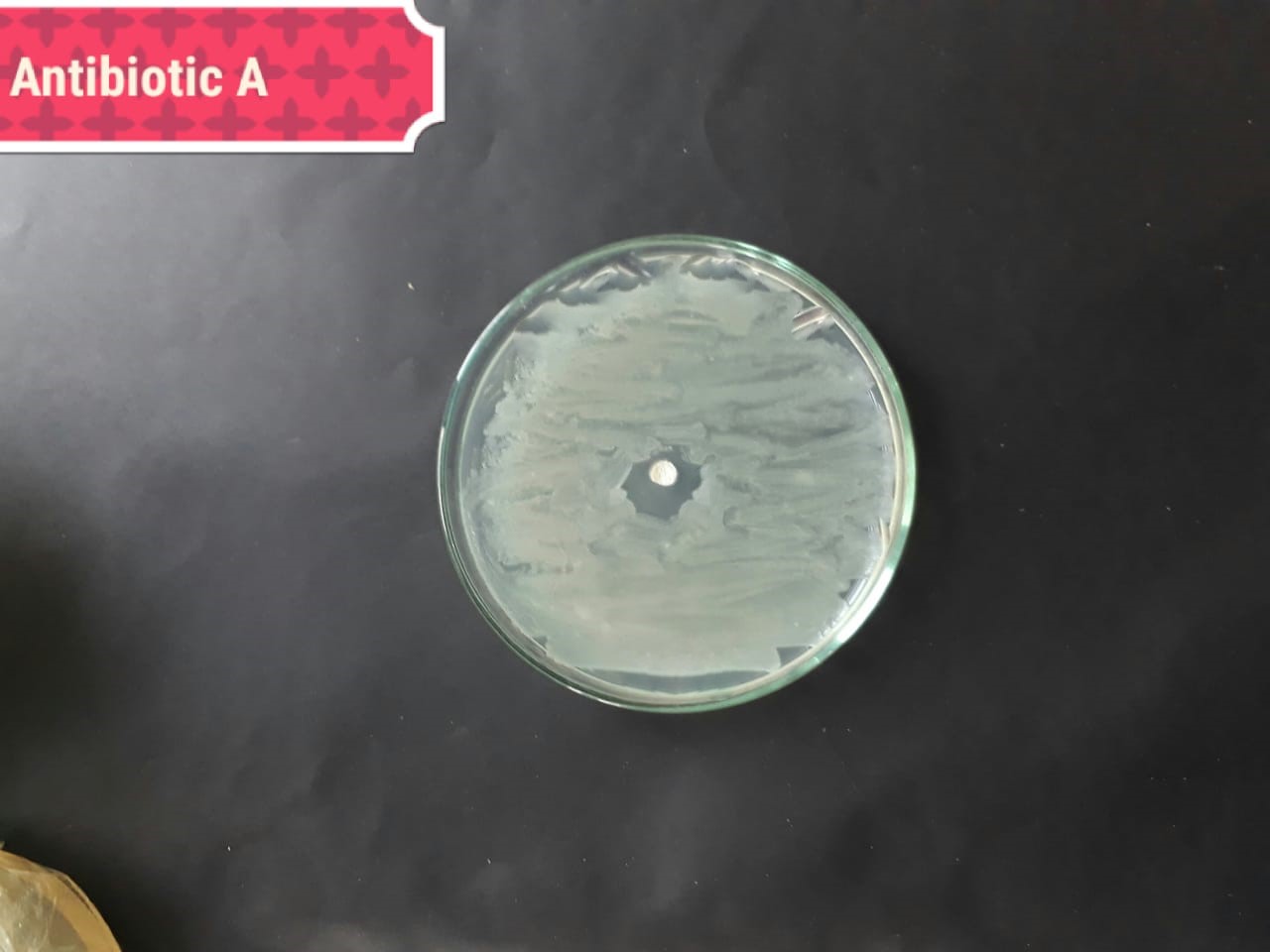 |
 |
| (MT416448) | 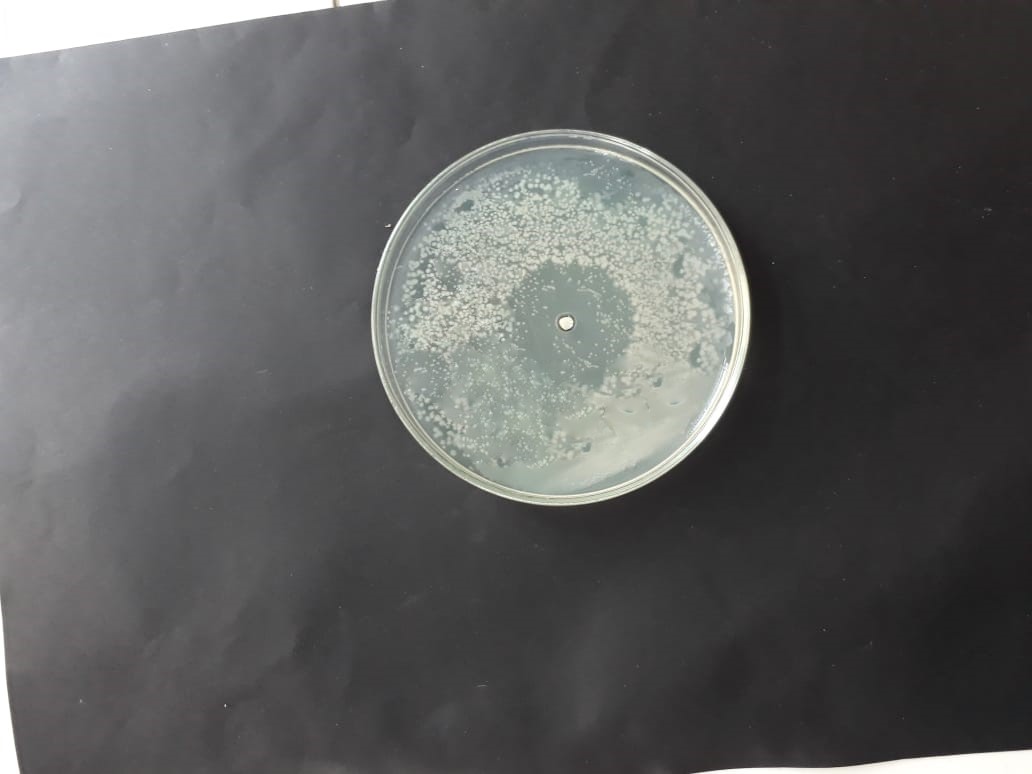 |
 |
| (MT416447) | 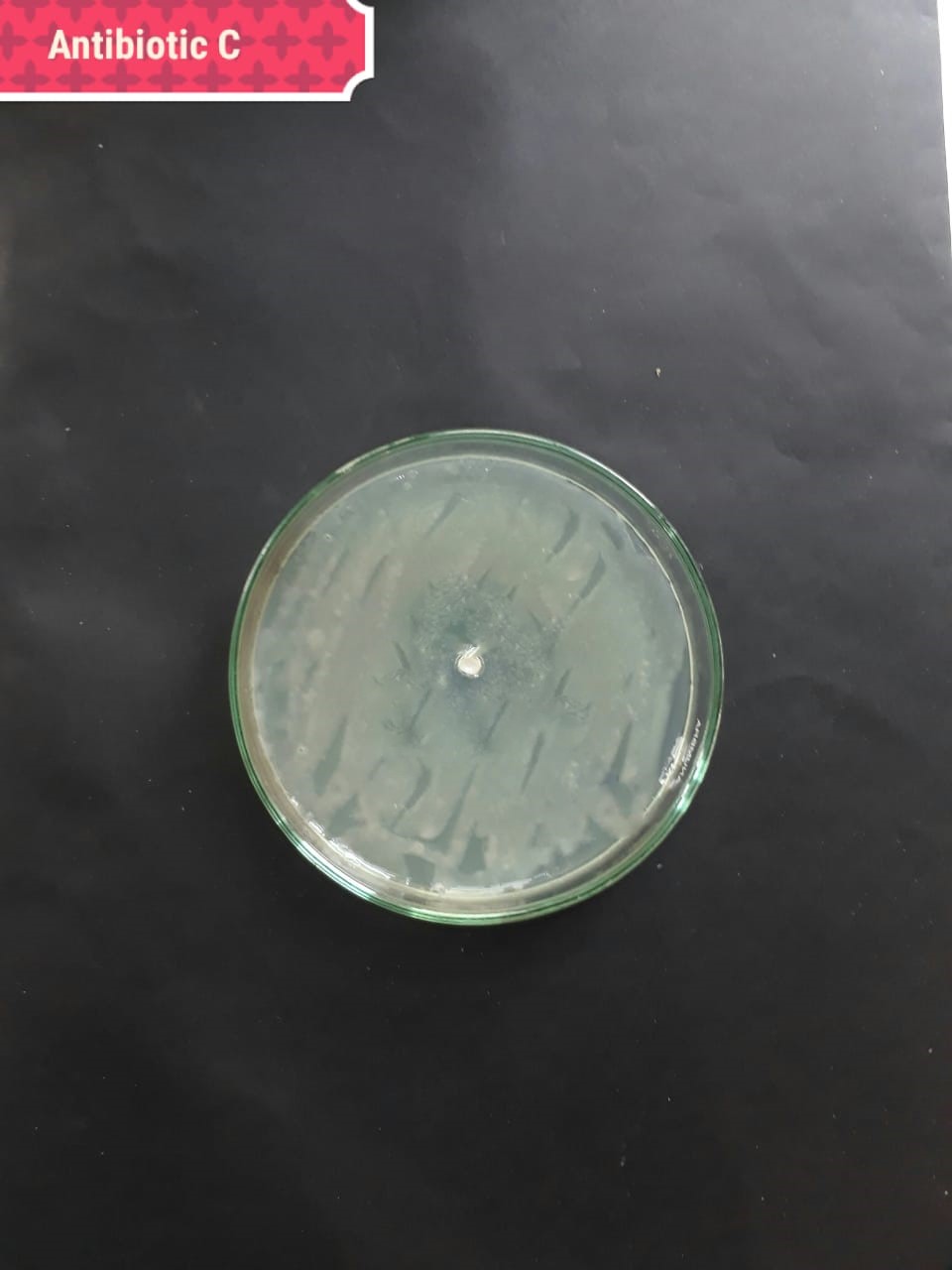 |
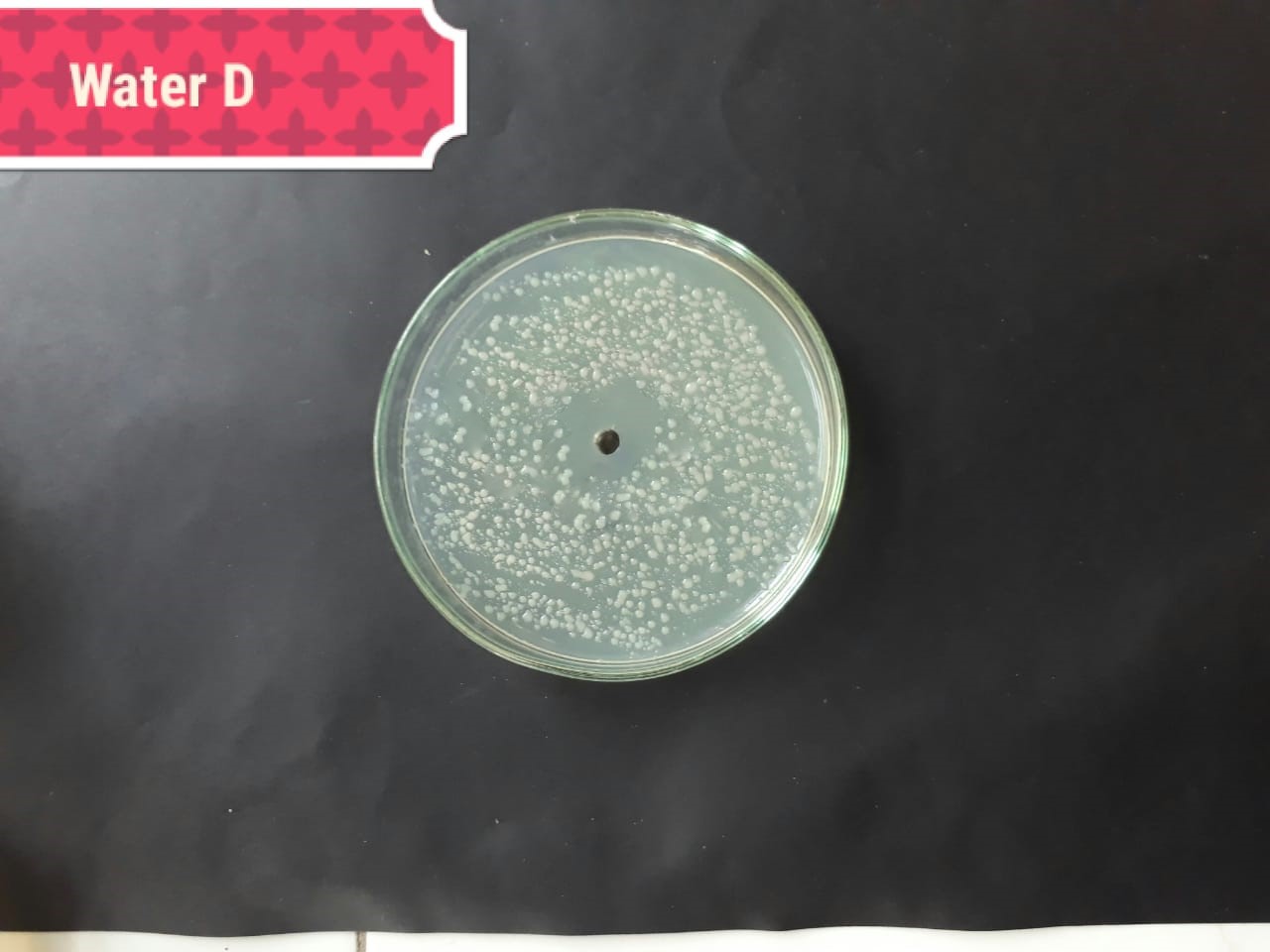 |
| (MT416446) | 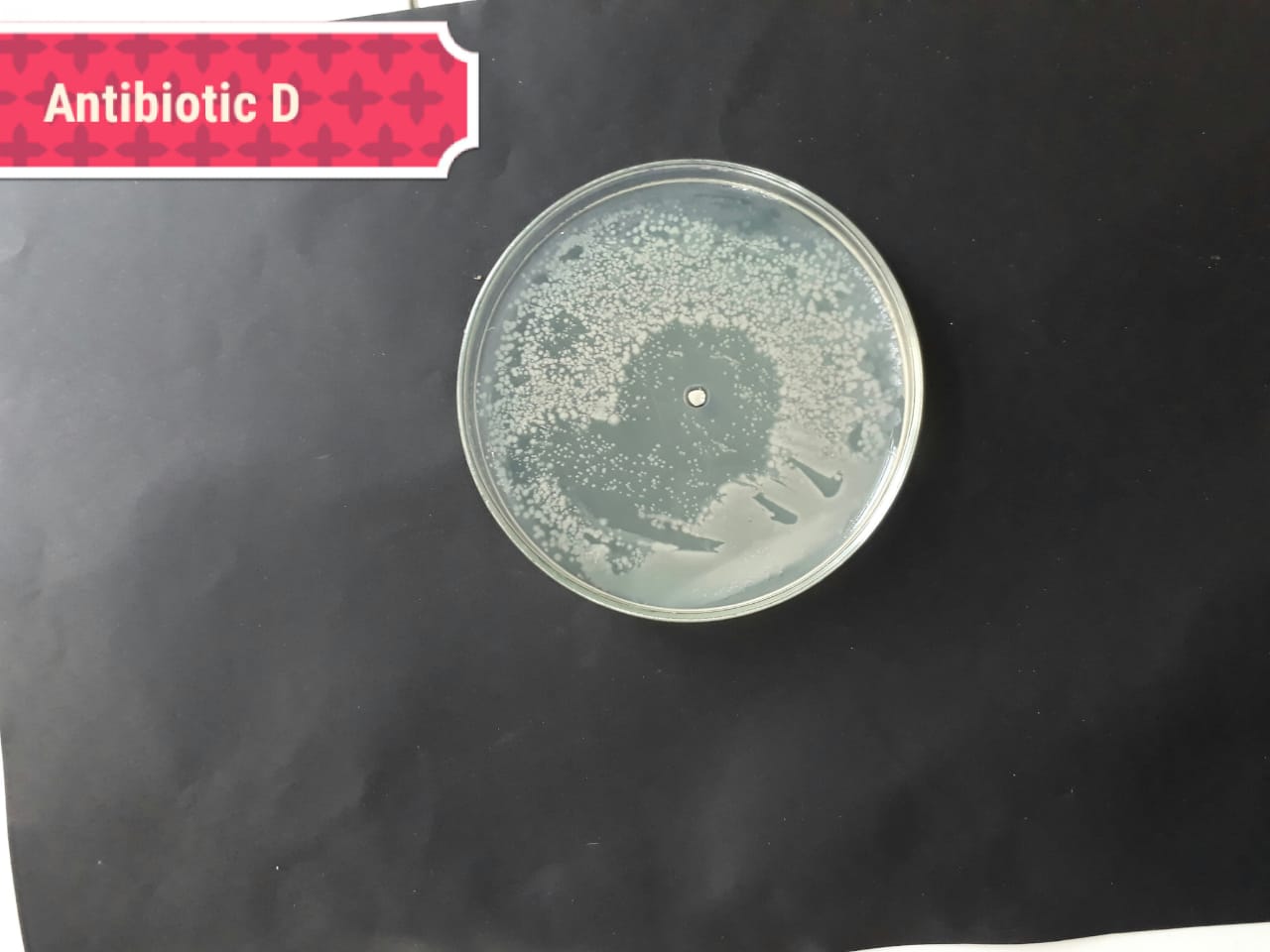 |
 |
The present investigation suggested high antibacterial activity of silver nanoparticles of H. porphyriformis against dental pathogens as compared to the particles of S. robusta. This is due to the smaller size of 15.23 nm with narrowest size distribution and spherical shape of the particles of H. porphyriformis, which makes them more stable [53, 70]. Smaller particles have high surface to volume ratio, so they release more silver ions and proved to destroy more bacteria actively [71]. These particles easily pass through the peptidoglycan layer of bacterial cell wall because of their smaller size and thus damage the respiratory chain by disrupting the respiratory chain reaction. These results are supported by previous research [72, 73]. The dental pathogens were tested againt silver nanoparticals synthesized from red algae using the zone of inhibition and minimum bacterial concentration. The larger ZOI were observed in low concentration than those reported by other studies [68, 69, 73, 74, 75]. Furthermore the low concentrations exhibit more therapeutic potential. It is further reported that the least MIC and MBC depends on the partical size and the aggregation of the NPs [76]. Furthermore ,previous studies also showed that antibacterial activities depends on dose [77, 78]. It is clear from Tables 2,4,5 that low concentarion of NPs causes high MIC and cell death which is thought to be due to the reaction between the NPs and bacterial proteins present in the cell wall [79, 80, 81].
The present study proposed improved characterization techniques due to emerging advancement in this field would expedite controlled, comparative and approximate performance of AgNPs. Present research concluded that silver nanoparticles synthesized by using red algae are cost effective and helpful antibacterial agents and can be used in the future against oral pathogens. The results of XRD and SEM confirmed the possible destructive interaction towards the bacterial cell wall. The antibacterial activity is attributed highly penetrated due to the smaller size of the green synthesized nanoparticles Above study in this document showed that, in the near future, an exceptional boom may be observed in the biosynthesis of algae-based NPs that will possibly have remarkable applications of antibacterial agents against the treatment of very resistant dental bacterial pathogens.
AgNPs, Silver Nanoparticles; FTIR, CFU, Colony-Forming Unit; Fourier-Transform Infrared Spectroscopy; MBC, Minimum Bactericidal Concentration; MIC, Minimum Inhibitory Concentration; NPs, Nanoparticles; SEM, Scanning Electron Microscopy; XRD, X-Ray Diffraction; ZOI, Zone of Inhibition.
The collection of samples and idea of research was given by UH and KDK. Bacterial strains were provided and identified by IL and SB. Statistical analysis and experimental process was guided by SS and UFA. Experimatation was done by the help of MDK and SI while formatting of manuscript was done by LP. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
Not applicable.
Not applicable.
This work had the support of “Fundação para a Ciência e Tecnologia (FCT)”, through the strategic projects (No. UID/MAR/04292/2020) granted to MARE.
The authors declare no conflict of interest. LP is serving as one of the Guest editors of this journal. We declare that LP had no involvement in the peer review of this article and has no access to information regarding its peer review. Full responsibility for the editorial process for this article was delegated to Baohong Zhang.