Circular RNAs (circRNAs) are a class of noncoding RNAs that are differentially expressed in tissues with a great potential in regulating the tumorigenesis and metastasis. The COX proportional hazard model predicts that among these, the relative expression of circRNA_104916 and lymphatic metastatic status independently predict the prognosis of patients with cancer. In the present study, we show that circRNA_104916 is downregulated in colorectal cancers as compared to the adjacent normal tissues. The expression of circRNA_104916 is negatively correlated with the tumor size, T stage and lymphatic metastasis. Patients with higher relative circRNA_104916 expression have a better post-operative disease-free survival as compared with those with lower relative expression. Forced expression of circRNA_104916 induces cell apoptosis and G2/M phase arrest by activating apoptosis pathway and cyclin proteins, thereby inhibiting proliferation of colon cancer cell lines, including LoVo and Caco-2. Additionally, overexpression of circRNA_104916 significantly suppresses migration and invasion of tumor cells by inhibiting the epithelial-mesenchymal transition. In conclusion, this study reveals that circRNA_104916 is a potential biomarker and therapeutic target for colorectal cancers.
Colorectal cancer (CRC) is the fourth most commonly diagnosed cancer and the second leading cause of cancer-related mortality worldwide according to the latest update of global cancer statistics (1). Surgery, including endoscopic resection, laparotomy and laparoscopic surgery, combined with adjuvant chemotherapy and neoadjuvant radiotherapy are the main therapeutic methods for CRC, however, the outcome of patients with CRC remains unsatisfactory (2). The 5-year survival rates markedly decline from about 90% in early-stage locally confined tumors to only about 5% in cases with metastatic disease, which is dependent on the tumor stage at a great extent (3). And local recurrence and distant metastasis, particularly within the digestive system are not uncommon among survivors (4). The molecular events that participated in the progression of CRC are worth further studies even if a considerable effort have been taken in the past years.
Circular RNAs (circRNAs), firstly identified in RNA virus and later observed in eukaryotic cells as an endogenous RNA splicing product, are a class of non-coding RNAs with covalently bonded circular structure, but without 5′ caps and 3′ tails (5, 6). Thanks to the development of high-throughput sequencing, molecular biology techniques and bioinformatics, the expression patterns and diverse functions of circRNAs are being elucidated. They are involved with multiple physiological and pathological processes through sponging miRNAs (7) and other RNAs (8) by base pairing, interacting with proteins by their three-dimensional structure (9) with a high degree of tissue-specific expression. Recently, a vast number of studies have revealed that circRNAs also play a key role in the progression of human cancers (10).
CircRNA_104916, generated from 9q33.3. and 651nt at length of mature transcript, was created with exon 1, exon 3, exon 4, exon 5, exon 6 and exon 8 of NEK6 via a non-canonical mode of RNA splicing (11). Besides, circRNA_104916 was significantly downregulated in gastric cancer tissues compared with adjacent normal tissues, forced expression of circRNA_104916 in gastric cancer cells suppresses migration, invasion and the process of epithelial-mesenchymal transition (EMT), which indicates that circRNA_104916 take a great role in gastric cancer progression. However, little is known of circRNA_104916 in the other cancers, including CRC. Herein, we determined circRNA_104916 expression in a cohort of treatment-naïve CRC patients before surgery, and the relationship between circRNA_104916 expression and clinicopathological characteristics, post-operative survival was studied. Additionally, the potential role of circRNA_104916 on tumor proliferation and metastasis were further explored by in vitro and in vivo experiments.
The human CRC samples, and their paired adjacent noncancerous specimen were collected from 116 patients who had undergone CRC surgical resection without receiving any other treatment before surgery and been pathologically diagnosed with CRC between 2009 and 2014. After excision, the tissues were quickly frozen and stored at -80°C. The Ethics Committee of Shantou University Medical College approved this study, and the informed consent form regarding specimen use for scientific research was signed by each patient.
The CRC cell lines including Caco-2, LoVo, SW480, HT-29 and HCT116, and the immortalized intestinal epithelial cell line called FHC, were obtained from the National Infrastructure of Cell Line Resource (Shanghai, China). Caco-2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) medium (Gibco) supplemented with 20% fetal bovine serum (FBS; Gibco), while other CRC cell lines and FHC cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, CA, USA) supplemented with 10% FBS (Gibco). The cells were maintained at 37°C in a humidified atmosphere with 5% CO2.
CircRNA_104916 (hsa_circRNA_104916) cDNA plasmid (pcDNA3.1.) was firstly developed by GENESEED (Guangzhou, China). Following that, the lentiviruses expressing circRNA_104916 (Lv_circRNA_104916) and negative control (Vector) were subsequently constructed by GENE (Shanghai, China). The LoVo and Caco-2 cells were collected, seeded, then transfected with circRNA_104916 lentiviruses. To date, the multiplicity of infection (MOI) is 50 for LoVo cells, while 100 for Caco-2 cells. After the transfection for 48 hours, the LoVo and Caco-2 cells were again cultured with free medium in the presence of 5 μg/mL polybrene. After 5 days, the stable transfected cells were collected, and applied for further study.
According to the manufacturer’s instruction, 500ng total RNA was reverse transcribed to cDNA with the PrimeScript RT Master Mix (TaKaRa, Dalian, China), following by total RNA was extracted from the frozen tissues and cell lines with Trizol reagent (TaKaRa). After that, the relative circRNA_104916 expression, normalized to expression of the endogenous control GAPDH, was determined by RT-qPCR assays using the SYBR Premix Ex Taq II Kit (TaKaRa) on the StepOnePlus system (Applied Biosystems, CA, USA). Fold changes were calculated using the 2−ΔΔCt method. The primer sequences of CircRNA_104916 is: forward: 5′-GCTCGGTGACCTTGGTCTGG-3′ and reverse: 5′-GCGTGTTGGGATGCCTCTGT-3′ and GAPDH – forward: 5′-TGGATCAGCAAGCAGGAGTA-3′ and reverse: 5′-TCGGCCACATTGTGAACTT-3′.
After the construction of circRNA_104916 up-regulated cells, we collected seed these stable type cell lines, seeded them into 96-well plates at density of 3,000 cells per well, and cultured them with free medium. Cell proliferation was detected at 24, 48, 72 and 96 hours using the Cell Counting Kit-8 (CCK-8) assay (Dojido, Tokyo, Japan) according to the manufacturer’s protocol. Briefly, 10 μL of CCK-8 solution was added to each well and incubated for 2 h at 37°C. The solution was then measured spectrophotometrically at 450 nm.
Female nude mice (age, 3–4 weeks) were bought from the Experimental Animal Centre of Guangdong Province, Guangzhou, China. The anti-growth effect of circRNA_104916 on CRC cell line was determined in vivo after received the approvement from the Ethics Committee of Shantou University. The transfected LoVo and Caco-2 cells were cultured, harvested and washed with PBS buffer, then subcutaneously injected into the flanks of the mice (5 × 106/200 μL for each nude mouse). The tumor volumes were measured and calculated every 4 days using the following formula: Volume = (length × width × width)/2, to determine the in vivo anti-growth effect of circRNA_104916 on CRC.
Cell cycle distribution and apoptotic were analyzed on a FACS Canto II flow cytometer (BD Biosciences) and detected using a cell cycle detection kit (Keygen, Nanjing, China) and the Annexin V-FITC/PI apoptosis detection kit (Keygen), respectively, as per the manufacturers’ instructions.
The cell migration abilities were evaluated by Transwell assays performed using Transwell chambers (0.8. μm; Corning, NY, USA) without Matrigel coating (Corning). The complete medium was added to the lower chambers, and 5 × 104 cells suspended in serum-free medium were then added into the upper compartments. After 24-h incubation, non-migrating cells were removed from the upper surface of the membranes, while the cells remaining on the other side were fixed and stained with 0.5.% crystal violet and digitally imaged. The cell invasion abilities were evaluated by Transwell assays performed using Transwell chambers (0.8. μm; Corning, NY, USA) without Matrigel coating (Corning). The complete medium was added to the lower chambers, and 5 × 104 cells suspended in serum-free medium were then added into the upper compartments. After 24-h incubation, non-invading cells were removed from the upper surface of the membranes, while the cells remaining on the other side were fixed and stained with Gemsa staining and digitally imaged.
Total proteins were extracted from cell lines by using radioimmunoprecipitation (RIPA; Beyotime, Shanghai, China) buffer supplemented with the protease inhibitor phenylmethanesulfonyl fluoride (PMSF; 1:100; Beyotime). After detecting the protein concentrations by using the bicinchoninic acid (BCA; Beyotime) method, equivalent total protein was separated by 10% sodium-dodecyl-sulfate–polyacrylamide-gel electrophoresis (SDS-PAGE; Fdbio Science, Hangzhou, China) and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, MA, USA). The membranes were then blocked with TBST solution containing 5% skimmed milk (Solarbio) for 1 h at room temperature and incubated with primary antibodies at 4°C overnight. The primary antibodies used (all from Proteintech; MA, USA) were as follows: anti-cyclin D1 rabbit polyclonal antibody (1:500), anti-cyclin B1 rabbit polyclonal antibody (1:1000), anti-E-cadherin rabbit polyclonal antibody (1:1000), anti-N-cadherin rabbit polyclonal antibody (1:1000), anti-Caspase-3 rabbit polyclonal antibody (1:2000), anti-Cleaved Caspase-3 rabbit polyclonal antibody (1:500), anti-Bax rabbit polyclonal antibody (1:500), anti-Bcl-2 rabbit polyclonal antibody (1:500), and anti-GAPDH rabbit polyclonal antibody (1:5000). Subsequently, the membranes were incubated with horseradish-peroxidase (HRP)–labeled goat anti-rabbit secondary antibody (1:10,000; Proteintech) for 1 h at room temperature; the protein bands were visualized with an ECL substrate kit (Fdbio Science).
Results have been presented as mean ± SEM. All statistical analyses were performed using the Pearson chi-squared test, two-tailed Student’s t-test, or analysis of variance (ANOVA), as appropriate, with either GraphPad Prism 7.0. (GraphPad Software, Inc, CA, USA) or IBM SPSS Statistics 20.0. (IBM, IL, USA)). Survival data were evaluated using the Kaplan-Meier method and log-rank test. P < 0.0.5 was considered statistically significant.
To elucidate the potential role of circRNA_104916 in CRC, we firstly determined the circRNA_104916 expression in 116 paired tumorous and normal tissues obtained from patients with CRC by RT-qPCR assay. The results showed that circRNA_104916 is significantly lower in tumorous tissues compared with adjacent normal tissues (Figure 1A), and of the 116 cases recruited in this cohort, 68.1.0% (79/116) of them displayed a decrease of the relative circRNA_104916 expression (Figure 1B), which indicates that circRNA_104916 might take a role in the progression of CRC.
 Figure 1
Figure 1Expression of circRNA_104916 in colorectal cancer and the correlation between its expression and clinicopathological parameters, as well as disease-free survival. (A) CircRNA_104916 was significantly downregulated in tumorous tissues compared with that in adjacent normal tissues detected using RT-qPCR assay (n=116). (B) Among the patients recruited, 68.1.0% (79/116) of them showed a decrease of circRNA_104916 in tumorous tissue than adjacent normal tissue (Log2 (2-ΔΔCT)). (C) Patients with higher circRNA_104916 expression (n=58) had a better disease-free survival than those with lower (n=58) revealed by Kaplan-Meier analysis. (D) The relative circRNA_104916 expression was lower in patients with T3 and T4 stage than those with T1 and T2 stage analyzed by Mann-Whitney U test. (E) The relative circRNA_104916 expression was lower in patients with lymphatic metastasis (N1 and N2) than those without lymphatic metastasis analyzed by Kruskal-Wallis test followed by Nemenyi test. (F) The relative circRNA_104916 expression was lower in patients with tumor diameters exceeded 5cm than those with less than 5cm analyzed by Mann-Whitney U test. (G) No significant difference of the relative circRNA_104916 expression was observed between patients with various differentiation degrees analyzed by Mann-Whitney U test. * P < 0.05, ** P < 0.01, *** P < 0.001, NS, not significant.
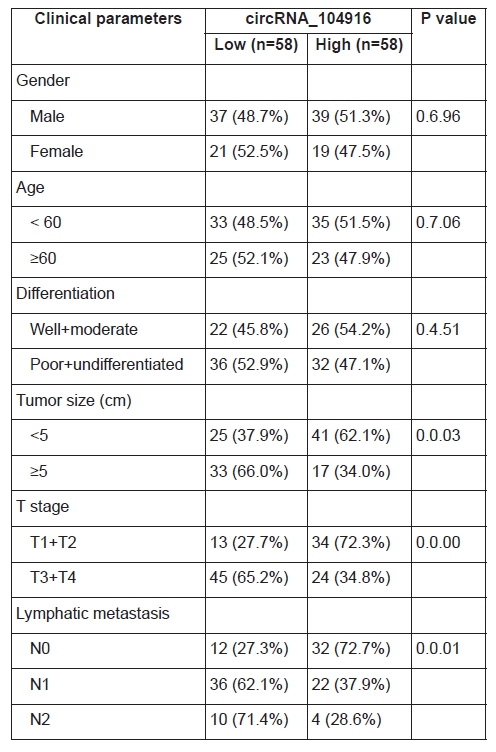
To evaluate the correlation between the relative circRNA_104916 expression and clinicopathological parameters, we divided the cohort into two groups as the median value (0.5.34). The chi-square tests (Table 1) showed that the relative circRNA_104916 expression was negatively correlated with tumor size (P = 0.0.03), T stage (P = 0.0.00) and lymphatic metastasis (P = 0.0.01), but no significant difference was observed between circRNA_104916 expression and other parameters (gender, age and differentiation degree). Furthermore, patients with higher degree of T staging (T1 + T2 versus T3 +T4; P <0.0.01; Figure 1D), those with lymphatic metastasis (N0 versus N1, P <0.0.5; N0 versus N2, P <0.0.1; Figure 1E), and those with larger tumor diameters (< 5cm versus ≥ 5cm, P < 0.0.5) inclined to have lower relative circRNA_104916 expression. And that patients in high circRNA_104916 group had a better performance for disease-free survival (DFS; could not determine the median value, yet) than those in low circRNA_104916 group (median, 865.0. ± 161.9. days) evaluated via Kaplan-Meier analysis (Figure 1C).
To measure the prognostic value of circRNA_104916 for DFS in patients with CRC, we established a COX proportional hazard model. Firstly, the univariate analysis showed that the relative circRNA_104916 expression, along with the differentiation degree, tumor size, T stage and lymphatic metastatic status, were significantly correlated with DFS in patients with CRC. Secondly, the multivariate analysis revealed that the relative circRNA_104916 expression and lymphatic metastatic status were able to independently predict patients’ prognosis (Table 2).

To further explore the potential role of circRNA_104916 in CRC, we conducted in vitro and in vivo experiments. To begin with, we determined the circRNA_104916 expression in five CRC cell lines and the immortalized intestinal epithelial cell line, FHC. The RT-qPCR analysis revealed that circRNA_104916 is also generally downregulated in CRC cell lines than that in FHC cell (Figure 2A). Based on this, we successfully overexpressed circRNA_104916 by a lentivirus-based method in Lovo and Caco-2 cell lines (Figure 2B and 2C), which expressing the lowest levels of circRNA_104916 in the cell lines displayed above (Figure 2A). After upregulation of circRNA_104916, the proliferation was significantly inhibited in both cell lines measured by CCK-8 assay (Figure 2D and 2E). In addition, the growth rate and weight of subcutaneously xenografted tumors in Lv_circRNA_104916 groups were also significantly suppressed than those of vector groups (Figure 2F and 2G). These results indicated that circRNA_104916 might function as a tumor suppressor gene in CRC progression.
 Figure 2
Figure 2Overexpression of circRNA_104916 inhibits proliferation of colorectal cancer cells in vitro and in vivo. (A) Expression of circRNA_104916 in a panel of colorectal cancer cell lines and the immortalized intestinal epithelial cell line, FHC, detected by RT-qPCR assay. (B and C) Successfully upregulation of circRNA_104916 by a lentivirus-based method in LoVo and Caco-2 cell lines. (D and E) The cell viability was significantly suppressed after overexpression of circRNA_104916 in LoVo (D) and Caco-2 (E) cell lines revealed by CCK-8 assay. (F and G) The growth rate and weight subcutaneously xenografted tumors in Lv_circRNA_104916 groups were suppressed than those of vector groups revealed by in vivo cell proliferation assay. * P < 0.05, ** P < 0.01, *** P < 0.001, NS, not significant.
The above in vitro and in vivo assays indicated that circRNA_104916 exerts an anti-growth effect in CRC cell lines, herein we intend to illuminate the impact of circRNA_104916 on cell apoptosis and cell cycle. The FACS showed that the apoptotic cells significantly increased after overexpressing circRNA_104916 (Figure 3A and 3B), at the same time, the content of cleaved caspase-3, apoptosis-related protein Bax were increased, while the anti-apoptosis marker Bcl-2 was inhibited in LoVo and Caco-2 cell lines (Figure 3C and 3D). For the alteration of cell cycle, the enhancement of G2/M phase cell rates was observed in Lv_circRNA_104916 groups measured by FACS (Figure 3E and 3F). The western blotting assay revealed that cyclin B1 was significantly upregulated, cyclin D1 was downregulated after overexpressing circRNA_104916 in both cell lines (Figure 3G and 3H). These in vitro assays indicated that circRNA_104916 inhibits cell proliferation through promoting apoptosis and G2/M phase arrest in CRC cell lines.
 Figure 3
Figure 3Upregulation of circRNA_104916 promotes apoptosis and the alteration of cell cycle in colorectal cancer cell lines. (A and B) The percentage of apoptotic cells was significantly increased after overexpression of circRNA_104916 in LoVo (A) and Caco-2 (B) determined by FACS. (C and D) The change of apoptosis-related protein expression (Caspase-3, cleaved Caspase-3, Bax), and the anti-apoptotic protein, Bcl-2 detected by western blotting. (E and F) The number of G2/M phase cells was significantly increased after overexpression of circRNA_104916 in in LoVo (E) and Caco-2 (F) determined by FACS. (G and H) The expression of G2/M phase checkpoint protein cyclin B1 was upregulated, while the G1 phase checkpoint protein cyclin D1 decreased when circRNA_104916 was overexpressed determined by western blotting. *** P < 0.001.
Transwell migration and invasion assays were conducted to evaluate the effect of circRNA_104916 on cellular migration and invasion capabilities. After overexpression of circRNA_104916, the number of migrated cells were significantly reduced in LoVo (Figure 4A) and Caco-2 (Figure 4B) cell lines. Similarly, the invasion assay showed the same results in LoVo (Figure 4C) and Caco-2 (Figure 4D) cell lines. To study the potential mechanism of circRNA_104916 on anti-migration and invasion abilities, the expression of EMT-related markers was detected by western blotting. E-cadherin, the epithelial marker, was upregulated, and the mesenchymal marker, N-cadherin was downregulated after overexpression of circRNA_104916 in both cell lines (Figure 4E and 4F).
 Figure 4
Figure 4CircRNA_104916 suppresses migration, invasion and process of epithelial–mesenchymal transition in colorectal cancer cell lines. (A and B) The number of migrated cells were significantly reduced in LoVo (A) and Caco-2 (B) cell lines measured by Transwell migration assay. (C and D) The number of invaded cells were significantly reduced in LoVo (C) and Caco-2 (D) cell lines measured by Transwell invasion assay. (E and F) The expression of the epithelial marker, E-cadherin (E-ca) was upregulated, and the mesenchymal marker, N-cadherin (N-ca) was downregulated after overexpression of circRNA_104916 in LoVo (E) and Caco-2 (F). *** P < 0.001.
CircRNAs have been implicated in cancer biology, as they are highly related with the occurrence of several human tumor types, even play important roles as cancer promoters or suppressors (12, 13). For example, circ-001569 is reported to promote tumorigenesis and invasion of hepatocellular carcinoma cells (14, 15), and circ_0066444 suppresses proliferation and metastasis of gastric cancer (16). Among all, low expression of circRNA_104916 has already been identified in gastric cancer, which is supposed to inhibit the metastasis of gastric cancer cells (11). However, little is known about circRNA_104916 in CRC.
In the present study, we explored the biological value of circRNA_104916, following by analyzing the expression pattern of circRNA_104916 in CRC tissues. CircRNA_104916 expression levels in CRC tissues were markedly lower than those in the corresponding noncancerous tissues. Moreover, the clinical parameters including tumor size, T stage, lymph node metastasis and poor prognosis, were found to be highly correlated with the downregulated circRNA_104916 expression in CRC patients. Consistently, the Cox analyses showed that lymph node metastasis and expression of circRNA_104916, were individual risk factors for disease-free survival in CRC patients. To date, Li’s study also reported that GC patients with invasion depth, higher tumor stage and more frequent lymphatic metastasis, had a lower expression of circRNA_104916 (17). Consequently, our findings consistently indicate that circRNA_104916 should be considered as a crucial molecular marker for CRC patients.
To elucidate the biological function of circRNA_104916 in CRC cells, in vitro and in vivo assays were performed by upregulating circRNA_104916 in the LoVo and Caco-2 CRC cell lines. We found that overexpression of circRNA_104916 could significantly suppress proliferation, and these results are in accordance with Li’s previous findings (17). Furthermore, our study also showed that overexpression of circRNA_104916 could dramatically induce apoptosis of CRC cells, and arrest CRC’s cell cycle in the G2/M phase.
Apoptosis is one form of programmed cell death, and Caspase-3-related pathways are the most common type of apoptosis (18). After being cell apoptosis triggered, cleaved caspase-3 could be dramatically upregulated, while the expressions of apoptosis markers are also changed. Furthermore, cancer cell growth is not only highly relevant with the apoptosis, but also with cycle progresses (19-21). Among all, Cyclin-pathway-related proteins are reported to be highly related with cancer cell cycle, whereas P53 plays an important role during the cell cycle process (22). Additionally, anti-apoptosis marker Bcl-2 could be transcriptionally depleted, whereas Bax is directly overexpressed, since the expression of p53 is activated (23, 24). Our study consistently showed that, after up-regulation of circRNA_104916, the apoptosis proteins including cleaved caspase-3 and Bax, were upregulated, while the anti-apoptosis marker Bcl-2, was inhibited. Furthermore, consistent with the flow cytometry results, the expression of the G2/M phase checkpoint protein cyclin B1 was upregulated, while the G1 phase checkpoint protein cyclin D1 decreased when circRNA_104916 was overexpressed. These results validated the hypothesis that circRNA_104916 may function as an anti-oncogene during the cell proliferation, apoptosis and cycle in CRC.
In addition, upregulation of circRNA_104916 remarkably weakened the migration and invasion capabilities of CRC cells, indicating that circRNA_104916 might inhibit metastasis in CRC. These findings coupled with our discovery of the negative relationship between tissue circRNA_104916 expression and lymph node metastasis suggested that upregulation of circRNA_104916 expression in CRC tissues might enhance distant metastasis and thus result in unfavorable outcomes in CRC patients. EMT, an important hallmark of cancer progression, involves transformation of epithelial cells to a mesenchymal cell phenotype via a pathophysiological mechanism (25, 26). The relationship between EMT and circRNAs in cancer metastasis has attracted considerable attention recently; most research has focused on the expression of the EMT-associated regulatory factor, including the epithelial marker E-cadherin, and the mesenchymal markers N-cadherin (11, 27). In the current study, we found that circRNA_104916 upregulation led to upregulation of the protein levels of E-cadherin and downregulation of N-cadherin in CRC cells, suggesting that absence of circRNA_104916 might reverse EMT process. To date, Li’s study also reported that circRNA_104916’s up-regulation could inhibit the migration and invasion of CRC cells, as the EMT process was altered. Therefore, we believe that circRNA_104916 could inhibit metastasis by activating EMT in CRC.
CircRNA_104916 is found to be downregulated in CRC tissues, while the low-positive patients showed poor prognosis. Overexpression of circRNA_104916 suppressed proliferation both in vitro and in vivo. With up-regulation, circRNA_104916 could induce more cell apoptosis and arrest more cells at G2/M phases, by activating apoptosis pathway and cyclin proteins. Furthermore, overexpression of circRNA_104916 inhibited migration and invasion of CRC cells by inhibiting their EMT process. Our study revealed that circRNA_104916 might be a potential biomarker and therapeutic target for CRC. However, there are still some limitations in our findings. Firstly, the downregulation of circRNA_104916 is not applied to demonstrate its function in CRC, owing to the endogenous low-expression of in CRC cells. Furthermore, though circRNA_104916 exhibit strong regulatory effects in CRC, further studies might be need to elucidate its miRNAs- related downstream.
Lianyong Min, Huaiming Wang contributed equally to this work. We thanks the Editage Company for the language revision service, and the authors declare that they have no competing interests.
Abbreviations: CRC: colorectal cancer; CircRNAs: circular RNAs; EMT: epithelial-mesenchymal transition; DMEM: Dulbecco’s modified Eagle’s medium; FBS: fetal bovine serum; RPMI: Roswell Park Memorial Institute; CCK-8: cell counting kit-8; RT-qPCR: real-time quantitative polymerase chain reaction; FACS: Flow cytometric analyses; PI: propidium iodide; FITC: fluorescein isothiocyanate; SEM: standard error of mean; ANOVA: analysis of variance; DFS: disease-free survival.
