Blood-testis barrier (BTB) that is constructed by testicular Sertoli cells (SCs) is essential for spermatogenesis. Krüppel-like factor 6 (Klf6), a nuclear transcription regulator, is reported to be associated with tight junction molecules of BTB between SCs during spermatogenesis; however, the specific regulatory role and mechanism of Klf6 in BTB regulation are still unknown. Here, we primarily confirmed the temporal and spatial expression patterns of Klf6 in mouse testes. Then, Klf6 was silenced in mouse cultured SCs using either Klf6-siRNA or Klf6-shRNA lentivirus. We mainly found that: (i) Klf6 was indispensable for the proliferative activity of mouse SCs; (ii) Klf6 regulated the integrity and permeability of BTB; (iii) Klf6 knockdown led to the significant upregulation of Zo-1, Claudin-11 and Vimentin, and downregulation of Claudin-3. Furthermore, Zo-1 and Claudin-3, participated in the tight junction remolding, were determined as targets of transcription factor Klf6 by luciferase assay. In summary, our findings suggest that Klf6 regulates the BTB assembly and disassembly via mainly targeting Zo-1 and Claudin-3 in mouse SCs.
Sertoli cells (SCs) are the somatic cells in the seminiferous tubules of the testis that are essential for spermatogenesis. Tight junctions between SCs form the blood-testis barrier (BTB) and control the nutrients and growth factor flowing to haploid germ cells, preventing from the interferences from immune system and foreign harmful substances. The formation and maintenance of BTB structure require many important adhesion and tight junction proteins, including Claudin-3, Claudin-11, Occludin, and zonula occludens-1 (Zo-1) (1). The molecular dynamics of the BTB proteins during spermatogenesis has been investigated in mice for a long time (2). Spermatogenesis is closely associated with the integrity and permeability of BTB. SCs secrete proteins that form the molecular basis for Sertoli-germ cell interactions, such as transport or bioprotective proteins, proteases, protease inhibitors, and glycoproteins (3, 4). For instance, impaired gap junction connexin43 in SCs leads to azoospermia (5).
BTB reconstruction and molecule variation occur when sperm are released into the lumen during spermatogenesis. Old BTB is disappeared and the new BTB is rebuilt. In this process, Claudin 3 is transiently incorporated into new tight junction and then replaced by Claudin 11 (1). Our previous study suggests that early spermatocytes seem to play a vital role in directing BTB assembly by regulating Claudin 3 in a spermatogonial stem cell transplantation assay (6). Gene expression profiles are revealed by RNA-Seq to explore the underlying mechanisms. Krüppel-like factor 6 (Klf6), a nuclear transcription factor, shows a dramatic change during the process of new BTB reconstruction (6). It remains unknown whether Klf6 is a potential transcription regulator driving the BTB reconstruction and spermatogenesis initiation.
Klf6 is universally expressed in mammalian tissues. It belongs to the Klf family involving a set of zinc finger transcription factors (7). Klf6 is widely recognized as a tumor suppressor gene and it has been well-studied in various cancers, regulating cell cycle, proliferation and apoptosis (8-10). In glioma endothelial cells, Klf6 is involved in miR-181a-mediated regulation of blood-tumor barrier integrity and permeability. Klf6 can directly target tight junction-related proteins, such as Zo-1, Occludin, and Claudin-5, via binding to their promoters (11). Since the blood-tumor barrier shares some similar physiological structure with BTB, it will be interesting to determine whether Klf6 is involved in BTB regulation.
In our previous study, we revealed the dynamic changes in tight junction molecules between SCs during different stages of spermatogenesis (6). Intriguingly, Klf6 expression in SCs exhibits a gradual decrease along with the differentiation of germ cells. However, the precise role of Klf6 on BTB structure and BTB-associated proteins remains largely unknown. Here, we show that Klf6 regulates the BTB assembly and disassembly via mainly targeting Zo-1 and Claudin-3 in mouse SCs. Our study provides new insights into the cellular mechanisms of spermatogenesis.
ICR mice were purchased from the Experimental Animal Center, Chinese Academy of Sciences (Beijing, China). All experimental protocols were reviewed and approved by the Animal Care and Use Committee of the Institute of Zoology, Chinese Academy of Science.
SCs were isolated from the testes of 21-day-old mice as previous reports (12, 13). After decapsulating the testes, the collected seminiferous tubules were pooled and washed for three times with phosphate buffer saline. The tubules were digested for cell suspension, mainly containing SCs and type A spermatogonia. Cell suspension was then cultured at 35 ℃ with 5 % CO2 incubator in Dulbecco modified Eagle medium/Ham F-12 (DMEM/F12) containing 10 % fetal calf serum, and 100 U/ml of penicillin. After 48 hr, cells were subjected to hypotonic treatment with 20 mM Tris (pH 7.4) to lyse the residual spermatogonia to obtain SCs.
Small interfering RNA (siRNA) molecules specifically targeting klf6 mRNA were purchased from GenePharma (Shanghai, China). The sequences of siRNAs for Klf6 and negative control were provided in Table 1. SCs were transfected with Klf6-targeted siRNA or negative control by lipofectamine RNAiMAX reagent (Invitrogen, CA, USA) following the manufacturer’s procedure at a final concentration of 0.1 μM. 72 hr after transfection, SCs were harvested for the mRNA quantification, Western blot, and immunofluorescence analysis.
| Name | Sequence 5’-3’ |
| Klf6-367 | GGAACGCUAUCUUCAGAGUTT; ACUCUGAAGAUAGCG UUCCTT |
| Klf6-614 | CCCACGACCAAAUUUACCUTT; AGGUAAA UUUGGUCGUGGGTT |
| Klf6-984 | GUGAUGAGUUGACCAGACATT; UCAAACAGCAUAUCAGGGCTT |
| Negative control | UUCUCCGAACGUGUCACGU TT; ACGUGACACGUUCGGAGAATT |
Short hairpin RNA (shRNA) molecules specifically targeting klf6 mRNA were further constructed to make stable and long-term silencing effect on Klf6 gene. The effective siRNA (Klf6-984) was cloned into the pENTR/U6 for packaging lentivirus (14). The titer of lentivirus harboring klf6-shRNA was determined as 109 IU/ml. SCs were infected with klf6-shRNA lentivirus and collected for further analysis after 72 hr.
Transfected SCs were collected and labeled with the DeadendTM Fluorometric TUNEL System (Promega, CA, USA) following the manufacturer’s protocol. Briefly, the cells were fixed with 4 % paraformaldehyde on ice for 20 min and then incubated with 70 % ice-cold ethanol for 4 hr. After washing, the cells were resuspended in equilibration buffer for 5 min. Cells were incubated with reaction buffer containing equilibration buffer, nucleotide xix, and rTdT enzyme at 37 oC for 60 min. The reaction was terminated with 20 mM EDTA, and cells were further stained with DAPI (Beyotime, Shanghai, China) and analyzed with the FACSCalibur system (BD Biosciences, CA, USA).
Total RNA was extracted from both testes and cell samples with RNAprep Pure Micro Kit (TianGen, Beijing, China). Reverse transcription of purified RNA was performed with Reverse Transcription System (Abm, Beijing, China) following the manufacturer’s protocol. PCR was performed with Qingke mix (Beijing, China) and products were separated by 1.2 % agarose gel electrophoresis. Quantitative RT-PCR was performed with Power SYBR Green Master Mix (Applied Biosystems, CA, USA) and analyzed by QuantStudioTM 12 K Flex Real-Time PCR System (BioRad, MA, USA). Primer pairs of selected genes were listed in Table 2.
| Gene | Forward primer (Sequence 5’-3’) | Reverse primer (Sequence 5’-3’) |
| Klf6 | TATCTTCAGGATGAGCCCTGCTAC | AGACTTCACCAATGGGATCAGAGG |
| Klf6-RT | ACCCGACATGGATGTGCTCCCAAT | GCAGGGCTCACTCTGAAGATA |
| Zo-1 | GCCCTCCGATCATTCC | TTAGACATTCGCTCTTCCTC |
| Occludin | AAGAAAGATGGATCGGTAT | GGAGGCATGTCCTGTG |
| Claudin-3 | ACCAACTGCGTACAAGACGAG | CAGAGCCGCCAACAGGAAA |
| Claudin-11 | ATGGTAGCCACTTGCCTTCAG | AGTTCGTCCATTTTTCGGCAG |
| Vimentin | GATCAGCTCACCAACGACAA | GCTTTCGGCTTCCTCTCTCT |
| Gapdh | TGTGTCCGTCGTGGATCTGA | CCTGCTTCACCACCTTCTTGA |
Proteins were collected from testes and SCs and then separated by 12 % SDS-PAGE and transferred to PVDF membranes (Millipore, CA, USA). PVDF membranes were blocked with 5 % skim milk for 1 hr and then incubated with primary antibodies at 4 oC overnight. Primary antibodies were listed as follows: Klf6 (1:1000, ab15580, Abcam), ZO-1 (1:1000, 339100, Invitrogen), Occludin (1:1000, ab31721, Abcam), claudin-3 (1:1000, 187340, Invitrogen), claudin-11 (1:1000, 364500, Invitrogen), vimentin (1:1000, 5741T, CST), and GAPDH (1:10000, MB001, Baode). The enzyme labeled secondary antibodies corresponding to the species of primary antibody were incubated with the PVDF membrane for 1 hr. Protein bands were developed with the Supersignal West Pico Chemiluminescent Substrate (Thermo Scientific, CA, USA), and detected using ChemiDoc XRS system with Image lab software (Bio-Rad, MA, USA).
Immunofluorescent staining was performed as our previous report (6). Briefly, testes were freshly collected, fixed in 4 % PFA and embedded in paraffin. The paraffin sections of 5 µm were obtained with a microtome. Sections were incubated with the primary antibody at 4°C overnight. Antibodies were obtained commercially as follows: Klf6 (1:100, ab15580, Abcam), Sox9 (1:200, AB5535, Millipore), Zo-1 (1:200, 339100, Invitrogen), Wt1 (1:200, ab89901, Abcam), vimentin (1:200, 5741T, CST). After washing, sections were treated with FITC or TRITC-conjugated secondary antibodies (Jackson ImmunoResearch, PA, USA) for 45 min. Nuclei were stained with DAPI (Beyotime, Shanghai, China). Images were acquired using a Nikon Eclipse 80i fluorescence microscope (Tokyo, Japan) with Nikon CCD camera.
SCs would assemble an intact epithelium with a functional tight junction barrier, which was able to resist the conductivity of electrical current delivered by a Millicell Electrical Resistance System (Millipore, MA, USA). Briefly, SCs were firstly plated on matrigel-coated bicameral units (diameter 12 mm; pore size 0.45 μm, effective surface area 0.6 cm2; Millipore) with the cell density of 1.0×106 cells/cm2. Each bicameral unit was placed inside the well of a 24-well dish with 0.5 ml DMEM/F12 each in the apical and the basal compartments. The resistance was then quantified as the trans-epithelial electrical resistance (TER) in ohms (Ω) by placing two electrodes across the SC epithelium, one in the apical and one in the basal compartment of the bicameral units.
Testes were firstly fixed with 2.5 % glutaraldehyde in 0.2 M cacodylate buffer overnight. Then, testes were cut into small pieces (1 mm3) and immersed in 1 % OsO4 in 0.2 M cacodylate buffer for 2 hr at 4°C. After fixation, ethanol gradient dehydration was performed and samples were embedded in resin. Ultrathin sections were obtained with ultramicrotome (Leica, Germany). Sections were then stained with uranyl acetate and lead citrate, and observed with a JEM-1400 transmission electron microscope (JEOL, Japan).
Kfl6 cDNA in eukaryotic expressing vector pFlag-CMV4 were obtained from Addgene (MA, USA). Promoters of claudin-3, vimentin and ZO-1 were amplified from mouse genomic DNA and then cloned into PGL4.17-Luciferase vector (Addgene, E6721). Klf6-expressing plasmids, promoter-luciferase plasmids, and internal control pRL-TK-Renilla (Promega, CA, USA) constructs were co-transfected into HEK293T cells on 96-well plates using lipofectamine 2000 (Invitrogen, CA, USA) following the manufacturer’s protocol. Cell extracts were prepared 48 hr after transfecting with the lysis buffer provided in the Dual-Luciferase Reporter Assay System kit (Vigorous Biotechnology, Beijing, China). Luciferase activity was measured on a Synergy Neo2 Multi-Mode Microplate Reader instrument (Bio-Tek, MA, USA) and the Renilla luciferase activity was applied to normalize the firefly luciferase activity.
All experiments were independently repeated at least three times. Data was presented as mean ± SEM (standard errors). Statistical analysis was performed with student’s t-test. All statistical tests were performed with SPSS 13.0 software (SPSS, CA, USA). *P<0.05 was considered as significance.
The expression profile of Klf6 in various tissues of mice was examined using RT-PCR. Klf6 mRNA was universally expressed in lung, kidney, skin, brain, testis, ovary, liver, and heart (Figure 1A). Furthermore, the expression of klf6 mRNA and protein was examined during testis development. Klf6 mRNA level in testes was gradually increased from 2 to 7 day-post-partum (dpp), while it was decreased from 7 dpp to 56 dpp (Figure 1B). Western blot analysis revealed that the Klf6 protein level in testes was increased from 6 to 56 dpp (Figure 1C). The expression of Klf6 in Sertoli cells and Leydig cells of adult mouse testes was confirmed by Western blot (Figure 1D) and immunofluorescent staining (Figure 1E). Collectively, transcription factor Klf6 was consistently expressed during testis development from neonate to adulthood in mice.
 Figure 1.
Figure 1.
Expression of Klf6 in mouse testes. (A) Klf6 mRNA in various tissues of mice was analyzed with RT-PCR. Klf6 mRNA (B) and protein (C) level during testis development, from birth to adulthood (0-56 dpp). Gapdh mRNA and Gapdh protein was applied as internal control. (D) Klf6 protein level in Leydig cells and Sertoli cells. (E) Klf6 in adult mouse testes was examined by immunofluorescence staining. DAPI indicated the nucleus. Scale bar, 100 μm.
Neighboring SCs form BTB structure to support testis development and spermatogenesis (1-3). Here, we isolated and cultured mouse primary SCs to explore the roles of Klf6 in vitro. Isolated cells were confirmed to be authentic Sertoli cells using specific Sertoli cell markers Sox9 and Wt1; as expected, they were expressed in the nucleus of >90% of isolated cells (Figure 2A, B). Furthermore, tight junction protein Zo-1 (6) was expressed in the cortical zone of neighboring SCs (Figure 2C). Vimentin, a cytoskeletal protein, was localized in the cytoskeleton of cultured SCs (Figure 2D). Notably, immunofluorescent staining indicated that Klf6 protein was localized in the nucleus of these primary SCs (Figure 2E), consistent with earlier reports (6, 13). We suggest that transcription factor Klf6 was expressed in the nucleus of SCs and hypothesize that Klf6 may play some roles in the transcription regulatory network within SCs.
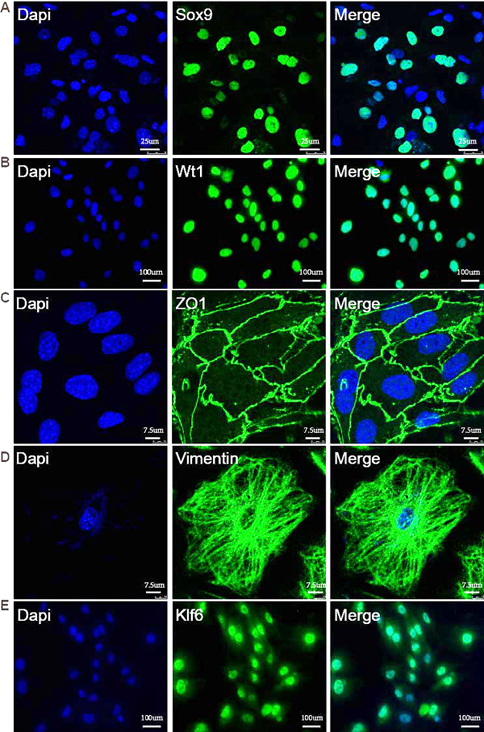 Figure 2.
Figure 2.
The expression of Klf6 in isolated SCs. (A, B) Isolated cells were characterized with SC specific markers Wt1 and Sox9 that were localized in the nucleus. (C) Immunostaining of tight junction marker Zo-1. (D) Immunostaining of cytoskeletal marker Vimentin. (E) Klf6 protein was localized in the nucleus of isolated primary SCs. Scale bar, 100 μm.
Klf6 plays important roles in the proliferation and differentiation of embryonic stem cells (15). To determine the potential roles of Klf6 in the regulation of SCs, Klf6 was knocked down with its specific siRNA duplexes and the proliferative activity was examined. After the transfection of klf6 siRNA, the number of SCs was considerably reduced as compared with negative control group (Figure 3A, B). RT-PCR and Western blot analyses revealed that Klf6 was successfully silenced by siRNA approach. The knockdown efficiency was ~50% (Figure 3C, D). Moreover, cell apoptosis was examined by TUNEL assay. The apoptosis of SCs was considerably increased by Klf6 knockdown (Figure 3E, F). Collectively, these data suggest that Klf6 was indispensable for the proliferation and cell viability of SCs.
 Figure 3.
Figure 3.
Knockdown of Klf6 and the proliferation and apoptosis of SCs. SCs were transfected with Klf6 siRNA or negative control siRNA for 72 hr. (A, B) Proliferation of SCs was reduced after transfection with Klf6 siRNA. (C, D) Klf6 mRNA and protein levels were quantified with quantitative RT-PCR and Western blot, respectively. Figure (E) was the statistical result of Figure (D). All the data were obtained from three independent experiments and analyzed by student’s t-test. **P<0.01, *P<0.05.
A lentiviral vector harboring Klf6-silenced shRNA was constructed, with higher transfection efficiency of approximately 70 % (Figure 4). SCs were infected with lentivirus and the integrity and permeability of BTB among neighboring SCs was further examined with electron microscopy. The typical BTB structure was observed among SCs infected with negative control lentivirus; however, BTB structure was not complete in klf6-shRNA lentivirus-infected SCs, exhibiting irregular cell edge (Figure 5A). SC tight junction permeability was examined daily by trans-epithelial electrical resistance assay (Figure 5B). Based on the resistance values (Ω), we found that the permeability of SCs tight junction was significantly disrupted after Klf6 silencing. Collectively, these data suggest that Klf6 played a critical role in the formation and/or maintain of tight junction and BTB structure of SCs.
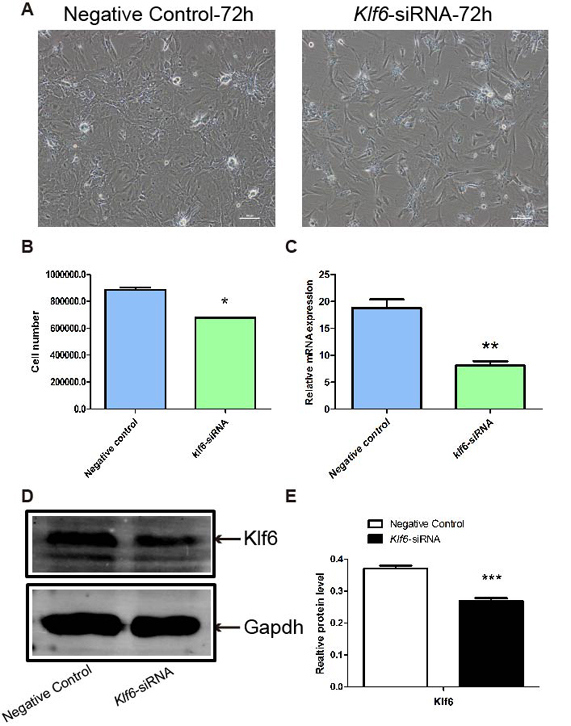 Figure 4.
Figure 4.
Knockdown of Klf6 by shRNA lentivirus infection. (A, B) SCs were infected with NC or Klf6-shRNA lentivirus for 72 hr. (C) Relative mRNA level of Klf6 was determined by quantitative RT-PCR. Gapdh mRNA was served as internal control. (D) Proliferation of SCs was reduced after transfection with Klf6 shRNA. Data was expressed as mean±SEM (n=3) and analyzed by student’s t-test. **P<0.01, *P<0.05.
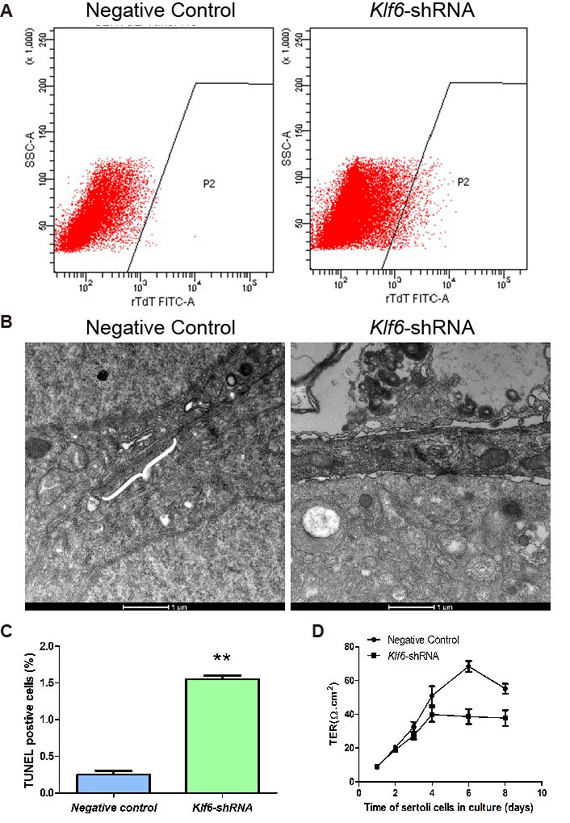 Figure 5.
Figure 5.
Klf6 regulated the integrity and permeability of BTB in SCs. (A) Apoptosis of SCs was increased after Klf6 silencing by flow cytometry. Figure (C) was the statistical result of Figure (A). (B) Electron microscopy assay indicated the complete BTB structure in the control group (White braces). The structure was disrupted in the Klf6-shRNA group and the edge of SCs appeared irregular. Scale bar, 1 μm. (D) The function of SC tight junction was examined daily by trans-epithelial electrical resistance (TER) from day 1 to 8 of culture. The solid circular was the negative control, and the solid cube was the Klf6-shRNA group, which was treated with Klf6 shRNA lentivirus in vitro. Data was expressed as mean±SEM (n=3) and analyzed by student’s t-test. **P<0.01, *P<0.05.
Loss of BTB integrity is always accompanied by disordered expression of BTB-associated proteins (e.g., adhesion proteins and tight junction molecules) (1-3). To explore the underlying mechanism of Klf6 regulation of BTB permeability, we examined the expression of BTB-associated markers, including Zo-1, Occludin, Claudin-3, Claudin-11, and Vimentin. The mRNA levels of Zo-1, Claudin-11, and Vimentin were significantly declined while Claudin-3 expression was significantly increased after Klf6 silencing. Occludin mRNA level was decreased in Klf6 knockdown group, while the result was not significant (Figure 6A). In consistent with RT-PCR results, Western blot analysis indicated that the protein levels of Zo-1, Claudin-11, and Vimentin were considerably decreased, and Claudin-3 protein was up-regulated (Figure 6B).
 Figure 6.
Figure 6.
Klf6 regulated the expression of BTB-related proteins in SCs. (A) After transfection with negative control lentivirus and Klf6-shRNA lentivirus for 72 hr, mRNA was extracted from SCs and quantitative RT-PCR was conducted for Zo-1, Occludin, Claudin-11, Claudin-3, and Vimentin. (B) Proteins were extracted and Western blot were performed for Zo-1, Occludin, Claudin-11, Claudin-3, and Vimentin. Gapdh mRNA and Gapdh protein were applied as internal control. Figure (C) was the statistical result of Figure (B). Data was expressed as mean±SEM (n=3) and analyzed by student’s t-test. ***P<0.001, **P<0.01, *P<0.05.
Candidate target genes of transcription factor Klf6 were determined with dual-luciferase assay. The binding motif sequence of Klf6 was identified and the potential Klf6 binding sites in promoter of Vimentin, Claudin-3 and Zo-1 were predicted with JASPAR (http://jaspar.genereg.net/) (16, 17). The predicted binding sites were further screened according to previous studies (11, 18, 19). It indicated that Klf6 bound to ‘CACCC’ and ‘GC box’ DNA sequences (Figure 7A, B).
 Figure 7.
Figure 7.
Identification of Klf6 target genes with dual-luciferase assay. (A, B) Scheme of the proximal promoters containing predicted Klf6-binding sites for Vimentin (from -519 to -40 of the TSS), Claudin-3 (from -566 to -141 of the TSS), and Zo-1 (from -700 to -1 of the TSS). Sequences of predicted Klf6 motif and potential Klf6-binding sites in Vimentin (M1, M2, M3), Claudin-3 (M4, M5, M6, M7), and Zo-1 (M8, M9, M10) (in red) were listed. TSS: transcription start site. (C-E) Dual-luciferase assay indicated the significant activation of Claudin3 and Zo-1 promoter by transcription factor Klf6. Data was expressed as mean±SEM (n=3) and analyzed by student’s t-test. *P<0.05.
The vectors of proximal promoters containing predicted Klf6-binding sites for Vimentin (M1, M2, M3; from -519 to -40 of the transcription start site), Claudin-3 (M4, M5, M6, M7; from -566 to -141 of the transcription start site), and Zo-1 (M8, M9, M10; from -700 to -1 of the transcription start site) were constructed. Klf6-expressing plasmids, promoter-luciferase plasmids, and pRL-TK-Renilla (internal control) constructs were co-transfected into HEK293T cells. We found that vimentin promoter luciferase activity was not obviously affected by Klf6 expression (Figure 7C). Notably, Klf6 significantly increased the luciferase activity of claudin-3 and ZO-1 promoters (Figure 7D, E). Collectively, transcription factor Klf6 may regulate the tight junction and BTB structure of SCs via targeting BTB protein Zo-1 and Claudin-3.
BTB is formed between SCs and consisted of tight junction strands, which restricts solutes, fluids and electrolytes from crossing the paracellular space, thus protecting meiotic and post-meiotic cells from impairment of immune system (1). Tight junctions of BTB form connection between neighboring cells, which are composed of several integral and peripheral membrane proteins, including Zo-1, Claudin-3, Claudin-5, Claudin-11 and Occludin (2). Notably, tight junctions are disassembled and reconstructed during the epithelial cycle of spermatogenesis. This is a complex process under precise regulation, involving the participation of various proteins and components (20). However, the underlying regulatory mechanisms on tight junction remolding and BTB assembly are still unclear.
Claudins family proteins are tight junction transmembrane proteins expressed in epithelia and endothelia, forming paracellular barriers that determine tight junction permeability. Claudin-3 is one of the well-studied tight junction proteins. Claudin-3 is expressed in seminiferous tubules, as a key constituent of the BTB (21). The permeability of BTB is controlled by androgen through regulating the expression of Claudin-3 (22). In the absence of androgen receptor, the expression of at least three tight junction protein components (e.g., Occludin, Claudin 11 and Claudin 3) of the BTB is reduced, leading to the increased permeability of BTB. Thus, androgen-induced variation of tight junction molecule expression is highly required for BTB remodeling (22).
Other TJ components involved in SC TJ remolding were also explored, such as Claudin-13 and a non-canonical TJ protein 2 isoform (23). Our previous study investigated the roles of germ cells in BTB assembly. Germ cells were capable of inducing Leydig cell testosterone production, which in turn enhance the expression of integral membrane protein Claudin-3 in SCs (6). Among different types of germ cells, early spermatocytes play a vital role in directing BTB assembly by regulating the expression of Claudin-3 in adjacent SCs (6, 23).
In glioma endothelial cells, Klf6 is involved in the miR-181a-mediated regulation of the blood-tumor barrier integrity and permeability via targeting the expression of Zo-1, Occludin, and Claudin-5 (11). Klf6 is specifically expressed in the nuclei of Leydig cells and SCs in adult mouse testes (15, 24). Knockdown of Klf6 in SCs impairs BTB integrity and dysregulation of BTB-associated proteins, including Zo-1, Claudin-11, Vimentin, and Claudin-3. Vimentin disruption was earlier shown to perturb barrier function of the BTB, as well as sperm quantity and quality (25). By utilizing luciferase assay, we further suggest that Klf6 regulates the BTB by targeting Zo-1 and Claudin-3. In Zo-1 deleted mouse testicular epithelial cells, the assembly of tight junctions is considerably prolonged; however, the final formation of tight junctions appears to be unaffected. Other member of the same protein family such as Zo-2 may compensate for the loss of Zo-1 (26).
Claudin-11, Occludin and Zo-1 are detected and distributed at (or near) the BTB throughout spermatogenesis, whereas claudin-3 is only expressed at the BTB at stage VI–IX of the epithelial cycle (2, 20). Tight junction remolding process, visualized by confocal microscope, further reveals that the differentiated germ cells migrate across BTB after old tight junctions are disintegrated. During this process, Claudin-3 is transiently incorporated into new tight junction to replace Claudin-11 (1). Variation tendency of key tight junction proteins in our study is consistent with the process of tight junction remodeling during germ cell migration. For instance, Claudin-11 expression is significantly declined and Claudin-3 expression is significantly increased after Klf6 silencing, which is accompanied with process of BTB breakdown. It remains to be determined the role of androgen in Klf6-mediated effects on BTB function. As we earlier reported that germ cells can up-regulate Leydig cell testosterone production (6), and testosterone was earlier shown to promote BTB function through its effects to induce BTB-associated protein production (e.g., Occludin, Zo-1) (27), perhaps via its effects by promoting protein recycling through intracellular protein trafficking (28). Thus, future studies should focus on the role of Klf6 in modulating steroidogenic pathway of testosterone production and/or androgen receptor homeostasis in the testis. Nonetheless, results of our study have unequivocally demonstrated that Klf6 is a novel regulator BTB remodeling in the testis. It is obvious that there are limitations in our study since only a few tight junction molecules functions through other pathways, which will be investigated in future studies. In addition, recent studies have shown the important roles of microRNAs in various biological processes. During BTB remodeling, it is possible that tight junction molecules are regulated by microRNAs through Klf6, which should be carefully evaluated in future studies.
This work was supported by grants from the National Key Research and Development Program of China (2018YFC1003500) and the National Natural Science Foundation of China (grants 31501953, 31471352, 31471400, 31501198, and 31171380). We also acknowledge support from the Clinical Capability Construction Project for Liaoning Provincial Hospitals (LNC CC-D50-2015 and LNC CC-C09-2015).
