- Academic Editor
†These authors contributed equally.
Background: Osteosarcoma cells are prone to metastasis, and the mechanism of N6-methyladenosine (m6A) methylation modification in this process is still unclear. Methylation modification of m6A plays an important role in the development of osteosarcoma, which is mainly due to abnormal expression of enzymes related to methylation modification of m6A, which in turn leads to changes in the methylation level of downstream target genes messenger RNA (mRNA) leading to tumor development. Methods: We analyzed the expression levels of m6A methylation modification-related enzyme genes in GSE12865 whole-genome sequencing data. And we used shRNA (short hairpin RNA) lentiviral interference to interfere with METTL3 (Methyltransferase 3) expression in osteosarcoma cells. We studied the cytological function of METTL3 by Cell Counting Kit-8 (CCK8), flow cytometry, migration and other experiments, and the molecular mechanism of METTL3 by RIP (RNA binding protein immunoprecipitation), Western blot and other experiments. Results: We found that METTL3 is abnormally highly expressed in osteosarcoma and interferes with METTL3 expression in osteosarcoma cells to inhibit metastasis, proliferation, and apoptosis of osteosarcoma cells. We subsequently found that METTL3 binds to the mRNA of CBX4 (chromobox homolog 4), a very important regulatory protein in osteosarcoma metastasis, and METTL3 regulates the mRNA and protein expression of CBX4. Further studies revealed that METTL3 inhibited metastasis of osteosarcoma cells by regulating CBX4. METTL3 has been found to be involved in osteosarcoma cells metastasis by CBX4 affecting the protein expression of matrix metalloproteinase 2 (MMP2), MMP9, E-Cadherin and N-Cadherin associated with osteosarcoma cells metastasis. Conclusions: These results suggest that the combined action of METTL3 and CBX4 plays an important role in the regulation of metastasis of osteosarcoma, and therefore, the METTL3-CBX4 axis pathway may be a new potential therapeutic target for osteosarcoma.
Osteosarcoma is a common primary bone malignancy that occurs mostly in adolescents and young adults with active skeletal development and is a malignant bone tumor with high capacities for invasion and metastasis [1, 2]. It usually arises in the distal femur and proximal tibia, and its pathogenesis is complex and closely related to genetic factors, environmental factors, and factors such as skeletal growth dysplasia [3, 4, 5].
Currently, the mainstay of treatment for osteosarcoma is a combination of surgical resection, chemotherapy, and radiation therapy, and the 5-year survival rate can exceed 60% [6].
However, due to the diversity and metastatic susceptibility of osteosarcoma, the
rates of recurrence and/or metastasis remain higher than 30%, and in affected
patients, the 5-year overall survival rate is
N6-methyladenosine (m6A) is a common chemical modification in eukaryotic messenger RNA (mRNA) [9]. This modification is widespread in eukaryotes and plays a critical role in regulating RNA processing, splicing, nucleation, translation, and stability, which are essential for the development of various human diseases, such as cancer [10, 11]. The effect of m6A modification on RNA depends mainly on the dynamic regulation of methyltransferases and demethylases [12]. It has been shown that the m6A level is high in osteosarcoma tissues and osteosarcoma cell lines and is associated with the malignancy of osteosarcoma [13, 14]. Another study found that m6A methylation regulated the expression of several key genes in osteosarcoma, including those associated with cell proliferation, invasion, and metastasis [15, 16]. These findings suggest that although the mechanism of metastasis in osteosarcoma cells has been investigated, studies on the mechanism of m6A methylation in osteosarcoma cell metastasis still need to continue.
Methyltransferase 3 (METTL3), an important m6A methyltransferase, is the only catalytic subunit mediating m6A methylation and plays a critical role in various cancer types [17]. In this study, we found that METTL3 was highly expressed in osteosarcoma tissues by analyzing whole-genome sequencing data in the GSE12865 dataset [18]. In further functional studies, we found that downregulation of METTL3 significantly increased the apoptosis and inhibited the proliferation and invasion of osteosarcoma cells. To explore the molecular mechanism by which METTL3 promotes metastasis in osteosarcoma, we examined CBX4 (chromobox homolog 4), a gene closely related to osteosarcoma cell metastasis [19] that is involved in the proliferation process in addition to the metastasis of osteosarcoma cells. METTL3 regulates the level of m6A methylation of CBX4 by binding directly to CBX4 mRNA, as demonstrated by the RIP (RNA binding protein immunoprecipitation) assay. Further experimental results showed that METTL3 could regulate the expression of CBX4 via m6A methylation of CBX4 and participate in the migration of osteosarcoma cells.
The human osteosarcoma cell lines MG-63 (FH0443, FuHeng Biology, Shanghai,
China) and HOS (Human osteosarcoma cells) (FH0440; FuHeng Biology) were cultured
in Dulbecco’s modified Eagle’s medium (DMEM; FHD01; FuHeng Biology) containing
10% fetal bovine serum (FBS, F8318, Sigma, USA) and 1% double antibiotic
solution (15140148, Gibco, USA). The cells were placed in a constant temperature
incubator at 37 ℃ for culturea nd 5% CO
Lentiviral vectors carrying a puromycin resistance cassette and a shRNA (short hairpin RNA) sequence targeting METTL3 or CBX4 were synthesized by Shanghai GenePharma (Shanghai, China), and the shRNA sequences are shown in Table 1. We transduced MG-63 cells and HOS cells with the METTL3 and CBX4 shRNA-containing lentiviral vectors separately. After 72 hours, we measured the RNA expression levels of METTL3 and CBX4 by qPCR and measured the protein expression levels of METTL3 and CBX4 by Western blotting. The CBX4 overexpression lentivirus was produced by Shanghai GenePharma. The overexpression efficiency of CBX4 was evaluated by both qPCR and Western blotting.
| Gene | Sequences (5 |
| shRNA-METTL3-1 | CGTCAGTATCTTGGGCAAGTT |
| shRNA-METTL3-2 | GCCTTAACATTGCCCACTGAT |
| shRNA-METTL3-3 | GCTGCACTTCAGACGAATTAT |
| shRNA-CBX4-1 | AGAGGCTGGTCGCCCAAATAT |
| shRNA-CBX4-2 | GCCCTTCTTTGGGAATATAAT |
Cells (HOS cells and MG-63 cells) from the experimental groups (shRNA-METTL3-1,
shRNA-METTL3-2) and the control group (shRNA-NC) in logarithmic growth phase were
seeded into 96-well plates at 1
Apoptosis was detected by flow cytometry using an Annexin V-FITC Apoptosis Assay
Kit (AT101, MultiSciences, Hangzhou, China). After removing the intervention
culture medium in each group of MG-63 cells and HOS cells (shRNA-NC,
shRNA-METTL3-1, shRNA-METTL3-2), trypsin was added to detach the cells, complete
DMEM culture medium was used to adjust the cell suspension concentration to
10
Total RNA was isolated using the MiniBEST Universal RNA Extraction Kit (9767,
Takara, Beijing, China) according to the manufacturer’s protocol, and total RNA
samples were reverse transcribed into cDNA using RT Master Mix (RR036A; Takara).
Real-time PCR was performed in triplicate using SYBR Green PCR Master Mix
(RR420A; Takara) and an ABI 7500 real-time PCR system. All procedures were
performed according to the manufacturer’s instructions. All primers were
synthesized by Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China) and
| Gene | Primer sequences (5 |
| METTL3 | Forward: ATCCCCAAGGCTTCAACCAG |
| Reverse: GCGAGTGCCAGGAGATAGTC | |
| CBX4 | Forward: TGAAATGGAGAGGCTGGTCG |
| Reverse: GCCCTCTCTTCCGATATCCC3 | |
| Forward: AGAGCTACGAGCTGCCTGAC | |
| Reverse: TAGTTTCGTGGATGCCACAGG |
Samples from each group were added to protein lysis buffer containing PMSF
(protease inhibitor), placed on ice for 40 min, and centrifuged at 12,000 r/min
for 40 min at 4 °C, and the supernatant was then collected. The protein
concentration in the supernatant was performed by the Bradford assay using BSA
(bovine serum albumin) as a standard. A 20 µg protein sample (20 µg)
was separated by 10% SDS‒PAGE and transferred to a PVDF (polyvinylidene
fluoride) membrane after electrophoretic separation, and the membrane was blocked
with blocking solution for 1 h, washed with PBS and incubated with primary
antibodies against METTL3 (DF12020, Affinity, OH, USA), CBX4 (DF13461, Affinity),
matrix metalloproteinase 2 (MMP2) (AF5330, Affinity), MMP9 (AF5228, Affinity),
E-Cadherin (AF0131, Affinity), and N-Cadherin (AF5239, Affinity) overnight at 4
°C. Following three washes in TBST, the membrane was incubated with
HRP-conjugated goat anti-mouse IgG (H + L) (SA00001-1, Proteintech, NY, USA) as
the secondary antibody for 1 h at room temperature. Bands were detected using a
gel documentation system. Band intensities were quantified using ImageJ software
(version1.46, U. S. National Institutes of Health, Bethesda, MD, USA), and all
values were compared to that of
MG-63 cells and HOS cells transfected with shRNAs were plated into the upper
chamber of 24-mm Transwell inserts (3412; Corning) (2
To verify the binding ability of CBX4 mRNA to METTL3, we performed RIP assays using the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (17-700, Millipore, MA, USA) according to a previously described method [20]. The antibodies used in the experiment were as follows: normal rabbit IgG (Cat# AC005, ABclonal, Woburn, MA, USA) and anti-METTL3 (Cat# A301-567A, Bethyl Laboratories, Montgomery, TX, USA).
The m6A immunoprecipitation (MeRIP) procedure was performed using a Magna MeRIP™ m6A kit (#17–10,499, Merck Millipore, MA, USA) according to instructions issued by the manufacturer. The steps are briefly described as follows: first, RNA was enriched and disrupted specifically; in the second step, RNA is incubated with antibodies (anti-m6A and IgG) and enriched specifically. We separated approximately 10% of the RNA in the m6A IP (immunoprecipitation) as input, and the third step was elution and reverse transcription–PCR. Next, we eluted the RNA from the anti-m6A antibody and IgG, as well as the input RNA, according to the requirements of the conventional kit, and performed reverse transcription with random primers. Through this experiment, we determined the effect of interference with METTL3 expression on the m6A level of cbx4 in HOS cells and MG-63 cells.
The experimental data are expressed as the means
We found that METTL3 was highly expressed in osteosarcoma tumor samples compared with normal human osteoblasts by analyzing the expression levels of genes encoding m6A methylation-related enzymes in whole-genome sequencing data from the GSE12865 dataset (Fig. 1A), and high METTL3 expression was negatively correlated with the survival of patients with sarcoma (data from GEPIA) (Fig. 1B).
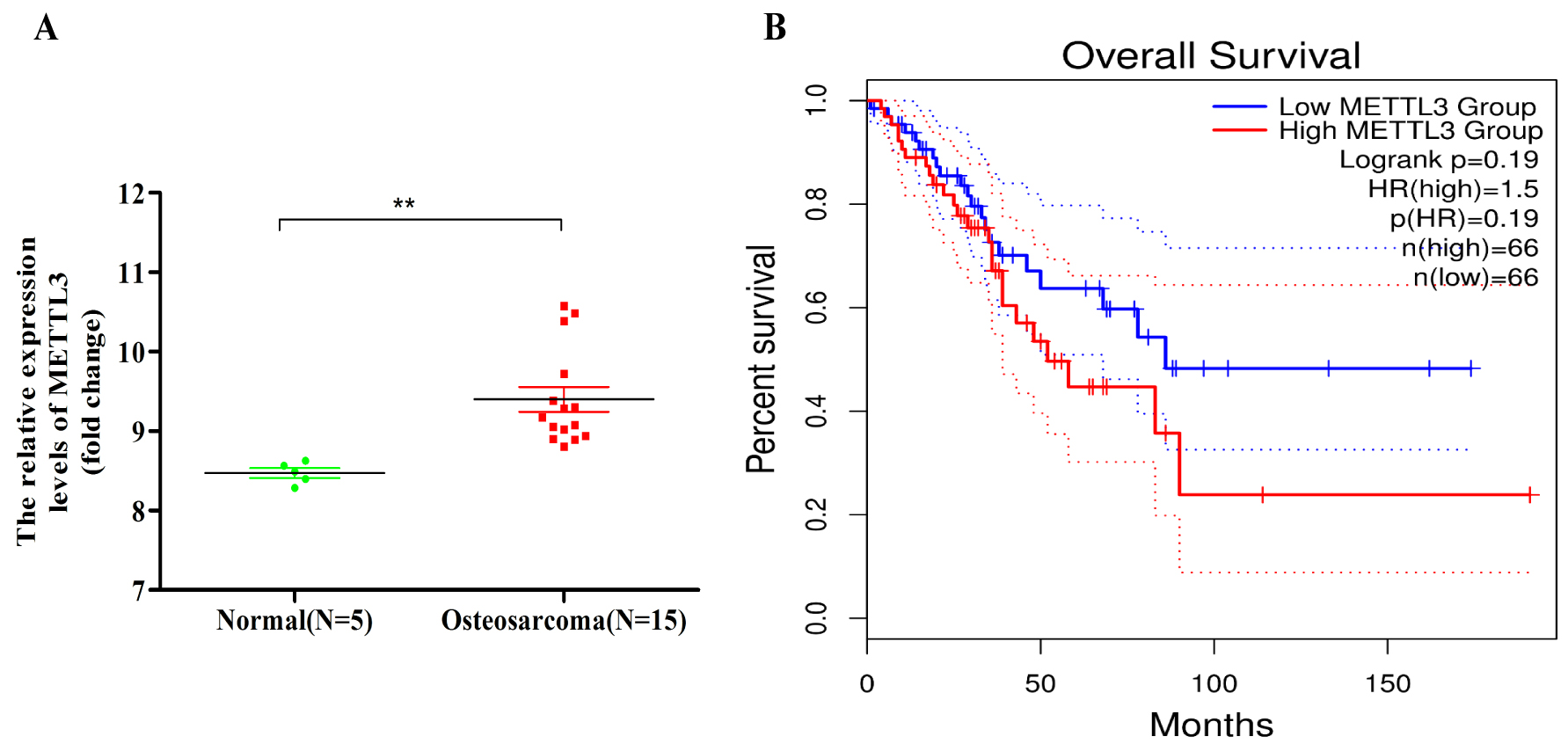 Fig. 1.
Fig. 1.METTL3 (Methyltransferase Like Protein 3) is highly
expressed in osteosarcoma and negatively correlated with the prognosis of
patients with sarcoma. (A) METTL3 was highly expressed in osteosarcoma tumor
samples relative to normal human osteoblasts, as determined by analyzing the
expression levels of genes encoding N6-methyladenosine (m6A) methylation-related
enzymes in whole-genome sequencing data from the GSE12865 dataset. **p
We then sought to further investigate the role of METTL3 in osteosarcoma cells. We used shRNA lentiviral vectors to interfere with METTL3 expression in osteosarcoma cells. We constructed lentiviral vectors containing three shRNAs targeting METTL3 to transduce HOS cells and MG-63 cells, and 72 hours later, we used RT‒PCR to determine the silencing efficiency of the three METTL3 shRNAs on METTL3 mRNA expression (Fig. 2A). Second, Western blotting was used to verify the effect of the three shRNAs on METTL3 protein expression (Fig. 2B). HOS cells and MG-63 cells were reinfected after screening for effective silencing of METTL3 by lentiviral shRNA transduction, and we evaluated the effect of METTL3 on cell metastasis (Fig. 2C). The results showed that among the three METTL3 shRNAs, shRNA-METTL3-1 and shRNA-METTL3-2 effectively inhibited the mRNA and protein expression of METTL3 and that silencing METTL3 significantly inhibited the metastasis of osteosarcoma cells.
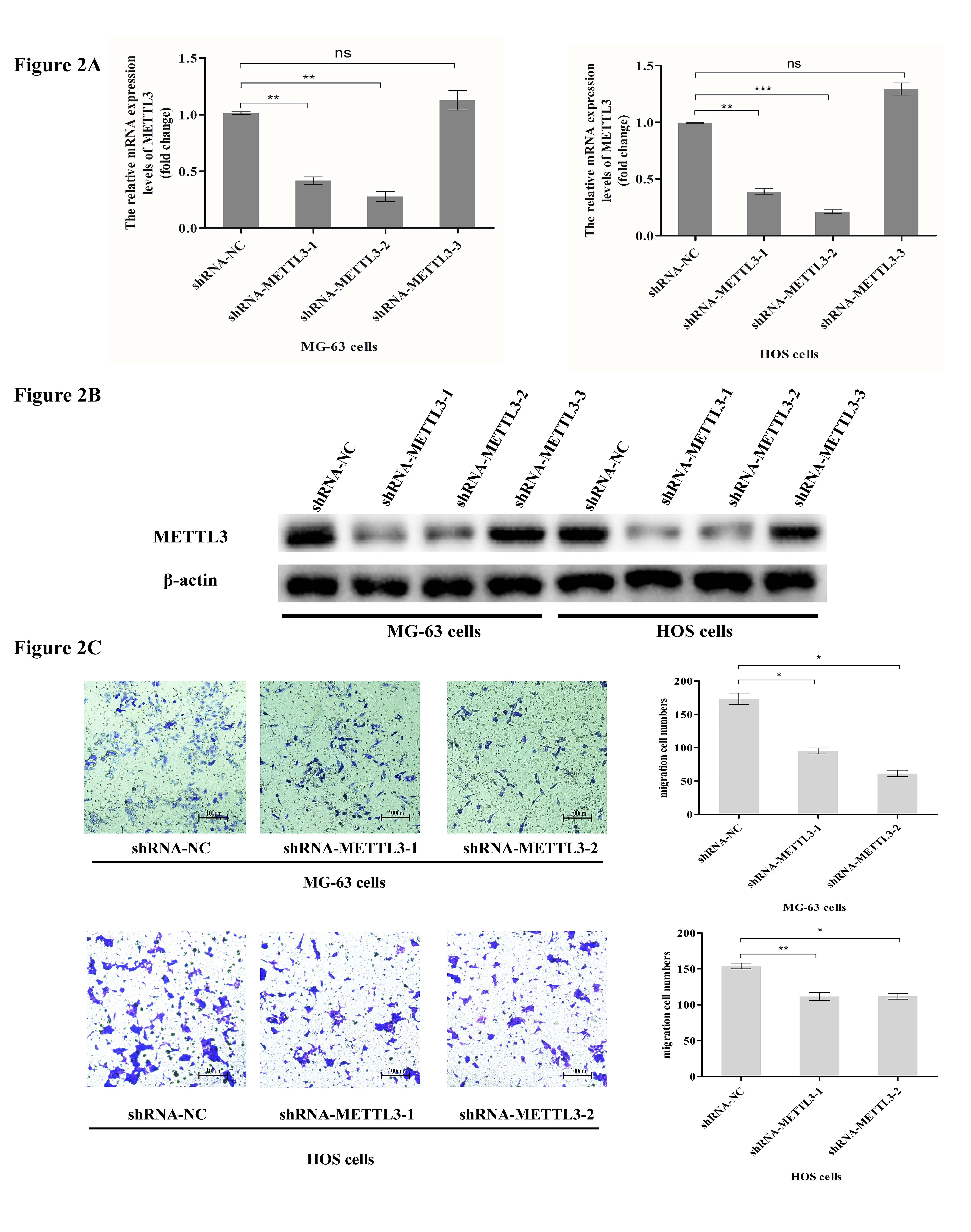 Fig. 2.
Fig. 2.METTL3 deficiency inhibits the metastasis of osteosarcoma
cells. (A,B) Lentiviral interference was used to silence METTL3 expression in
HOS cells and MG-63 cells. We used real-time PCR and Western blotting to
determine the silencing efficiency of METTL3. As shown in the figure, METTL3 RNA
and protein expression was significantly inhibited. (C) The migration ability was
determined using the indicated stable cells as described in the Methods section.
The results showed that silencing METTL3 significantly inhibited the metastasis
of osteosarcoma cells. The results are expressed as the mean
To further verify the effect of METTL3 on the biological functions of osteosarcoma cells, we examined the effect of inhibiting METTL3 expression on osteosarcoma cell proliferation by a CCK8 assay (Fig. 3A) and the effect of inhibiting METTL3 expression on osteosarcoma cell apoptosis by flow cytometry (Fig. 3B). The results showed that inhibiting METTL3 expression significantly inhibited the proliferation and promoted the apoptosis of osteosarcoma cells.
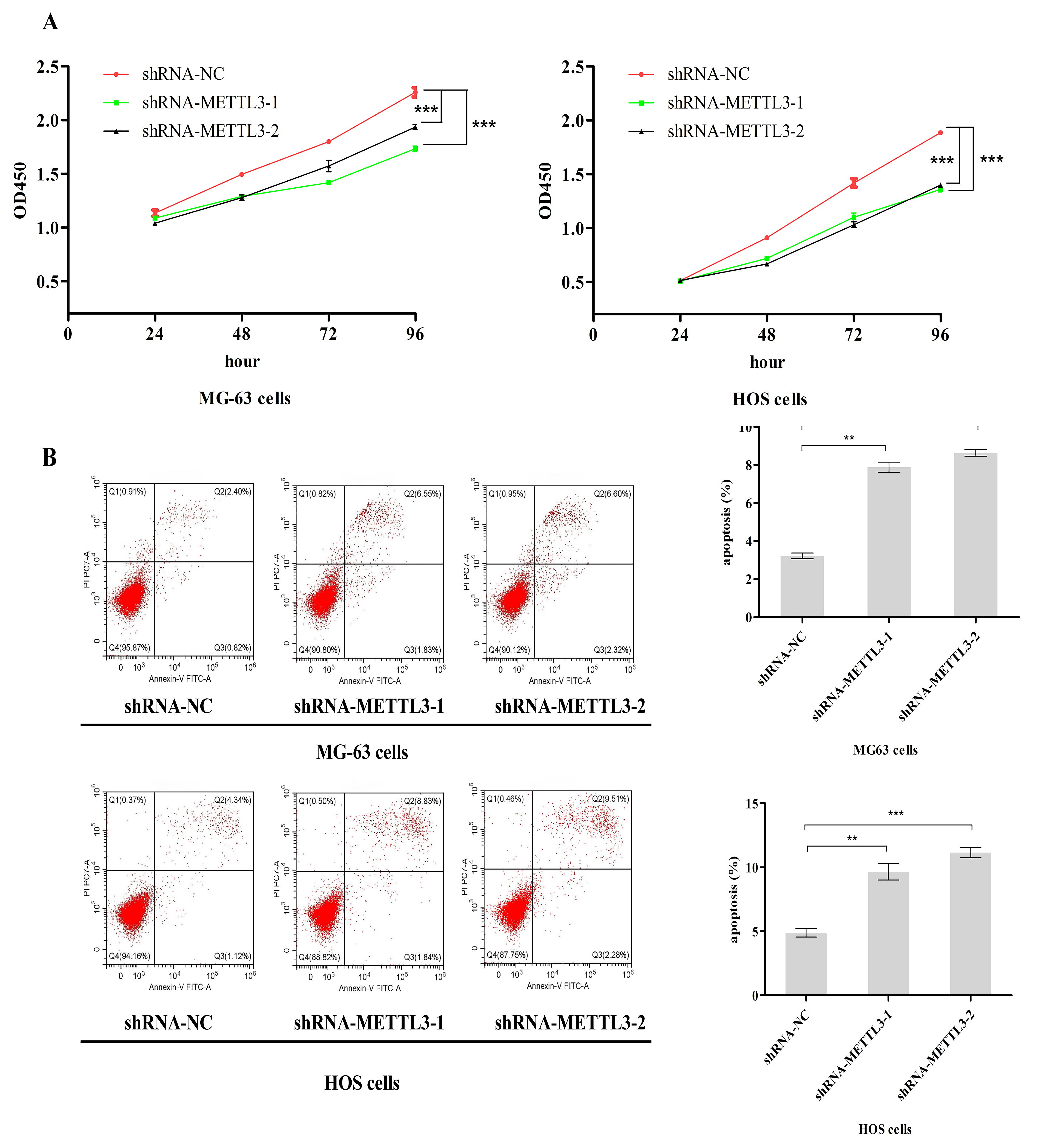 Fig. 3.
Fig. 3.METTL3 deficiency inhibits the proliferation and promotes the
apoptosis of osteosarcoma cells. (A,B) The proliferation and apoptosis abilities
were evaluated using the indicated stable cells as described in the Methods
section (Cell Counting Kit-8 (CCK8) assay and flow cytometric analysis of
apoptosis, respectively). The results showed that silencing METTL3 significantly
inhibited the proliferation and promoted the apoptosis of osteosarcoma cells (HOS
cells and MG-63 cells). The results are expressed as the mean
Previous literature reported that CBX4 transcriptionally upregulates Runx2 by recruiting GCN5 to the Runx2 promoter, thereby promoting the metastasis of osteosarcoma cells [19]. Our analysis of potential target genes downstream of METTL3 (data not shown) revealed that CBX4 was a potential target gene of METTL3; we then predicted through the SRAMP prediction server that CBX4 RNA contains m6A sites [21] (Fig. 4A), and detailed information about the predicted sites is shown in Table 3. We also detected the binding of METTL3 to CBX4 mRNA using RIP-qPCR (Fig. 4B). We demonstrated that METTL3 downregulated CBX4 RNA expression after interfering with METTL3 expression in HOS cells and MG-63 cells (Fig. 4C). These results demonstrated that there is a regulatory relationship between METTL3 and CBX4. Through MeRIP-qPCR, we found that silencing METTL3 significantly reduced the methylation level of CBX4 RNA in HOS cells and MG-63 cells (Fig. 4D).
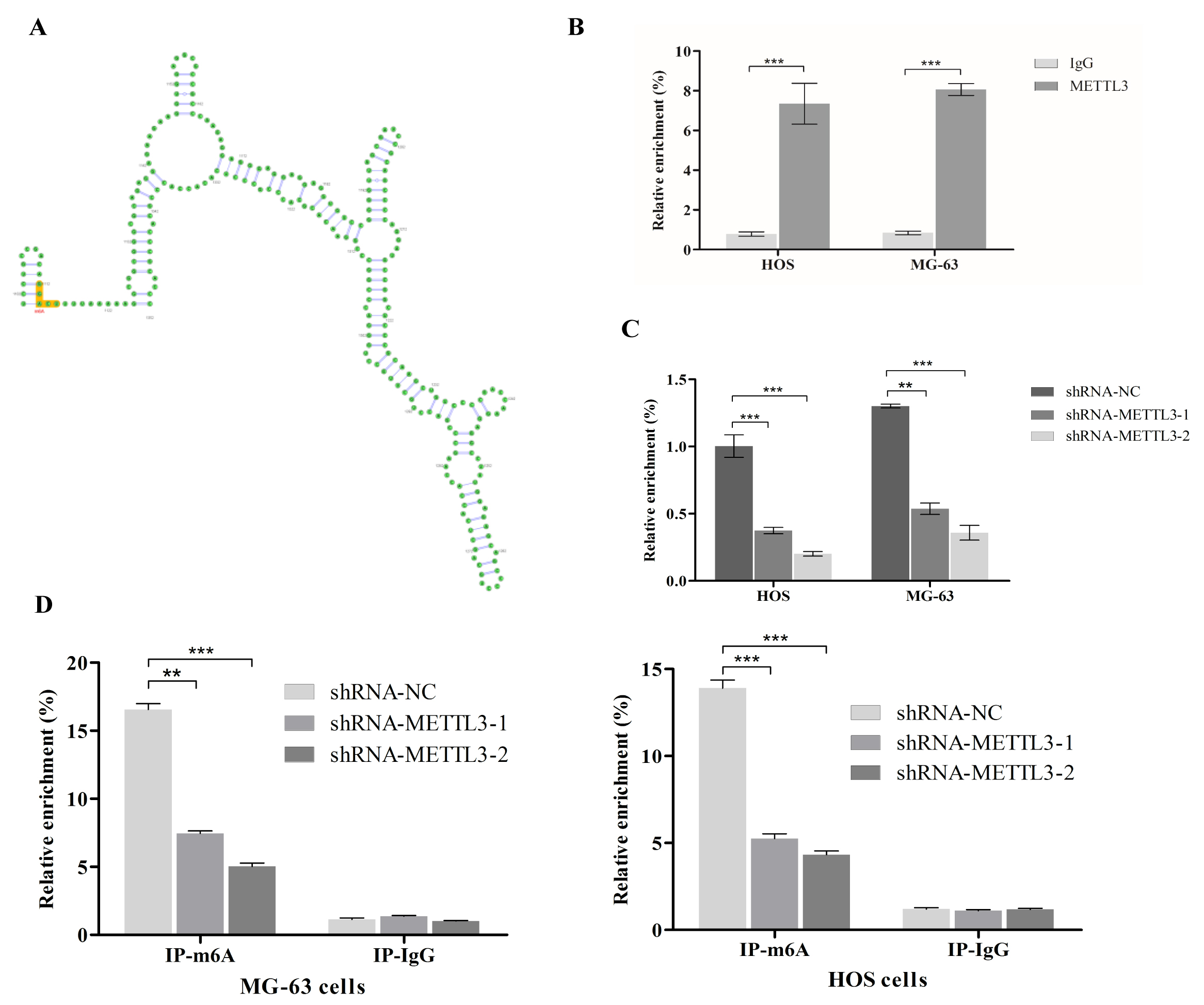 Fig. 4.
Fig. 4.CBX4 is regulated by METTL3. (A) CBX4 messenger RNA (mRNA)
contains m6A methylation sites according to the SRAMP prediction server (a
representative site map). (B) The RNA Binding Protein Immunoprecipitation (RIP) assay of METTL3 showed direct binding
between the METTL3 protein and the mRNA of CBX4 (three independent experiments,
***p
| # | Position | Sequence context | Structural context | Score | Score | Score | Score |
| (binary) | (knn) | (spectru) | (combined) | ||||
| 1 | 749 | GCGCC CAGCC CCACC | PPPPP PMPPP PPPPI | 0.812 | 0.648 | 0.842 | 0.815 |
| UGCCC GGACC UGGGG | PPIIP PPIII PPPPP | ||||||
| GCCAA GAGCC ACCCG | PPHHH HHPPP BPPPP | ||||||
| 2 | 1114 | GCGCC ACCCU CCUGC | PPPMM MMMMP PPPPH | 0.929 | 0.811 | 0.811 | 0.876 |
| CGACA GGACU UUUAA | HHHPP PPPMM MMMMM | ||||||
| AAAGG CGGCG GGCGC | MMMPP IIPPP PPIPP | ||||||
| 3 | 1586 | CAGCC CGAGG UCAUC | BBPPP PPIIP PPBPP | 0.831 | 0.644 | 0.881 | 0.841 |
| CUGCU AGACU CAGAC | IIPPP BBBBP PBBPP | ||||||
| CUGGA UGAAC CCAUA | PPPPP MMMMM MMMMM | ||||||
| 4 | 1592 | GAGGU CAUCC UGCUA | PIIPP PBPPI IPPPB | 0.725 | 0.357 | 0.864 | 0.762 |
| GACUC AGACC UGGAU | BBBPP BBPPP PPPPM | ||||||
| GAACC CAUAG ACUUG | MMMMM MMMMP IPPPB | ||||||
| 5 | 1602 | UGCUA GACUC AGACC | IPPPB BBBPP BBPPP | 0.74 | 0.596 | 0.863 | 0.782 |
| UGGAU GAACC CAUAG | PPPPM MMMMM MMMMP | ||||||
| ACUUG CGCUG CGUCA | IPPPB PPPPP PPPHH | ||||||
| 6 | 1610 | UCAGA CCUGG AUGAA | PPBBP PPPPP PMMMM | 0.865 | 0.65 | 0.821 | 0.837 |
| CCCAU AGACU UGCGC | MMMMM MPIPP PBPPP | ||||||
| UGCGU CAAGA CGCGC | PPPPP HHHHH HPPPP | ||||||
| 7 | 1832 | AUUAU CACCG ACGUC | MMMMM MMPPP PPPPP | 0.882 | 0.596 | 0.827 | 0.846 |
| ACCGC GAACU GCCUC | IPPPB BBBBB PPIIP | ||||||
| ACCGU UACUU UCAAG | PPPPP PPPHH HHHHH | ||||||
| 8 | 2136 | UGGAG AUGGA CCCCC | PPPIP HHHHH HPIPP | 0.855 | 0.437 | 0.804 | 0.814 |
| GGAAC GGACA GGGCA | PPIII PPPIP PPPPM | ||||||
| GCUCU GCGCC CGGCC | PPPPI PPIPP MPPPP | ||||||
| 9 | 2207 | CGUGC UAACU UGUCA | MMMMM MMMMMMPPPP | 0.799 | 0.424 | 0.752 | 0.761 |
| AGUGC UGACU CUACU | HHHHH PPPPM MMPPI | ||||||
| CCCGU UUGUA CGUGG | PPPPP HHHHP PPBPP | ||||||
| 10 | 2468 | CCCCU CUCUU CCCCU | IPPMM MMMMMMMMMM | 0.856 | 0.418 | 0.75 | 0.792 |
| UCCAC GAACU GCAAU | MMMMM MMMMM MMMPP | ||||||
| ACCAG UAACC UUGGU | PPPPP HHHHH PPPPP | ||||||
| 11 | 2589 | AGGCU ACACG AGCUC | MPPPP IIIIP PPPPP | 0.819 | 0.502 | 0.803 | 0.797 |
| UCUAG AAACU GCUGC | MMMMM MMMPP PPPPP | ||||||
| UACUA GAAAU GUCUA | PPPPM MMMMMMMMMM |
We used lentiviral shRNA transduction to interfere with CBX4 expression in osteosarcoma cells, and Fig. 5A,B show the changes in the RNA and protein expression levels after interference with CBX4 in HOS cells and MG-63 cells. The transwell assay data in Fig. 5C demonstrate that interference with CBX4 expression alone in HOS cells and MG-63 cells also inhibited osteosarcoma cell metastasis.
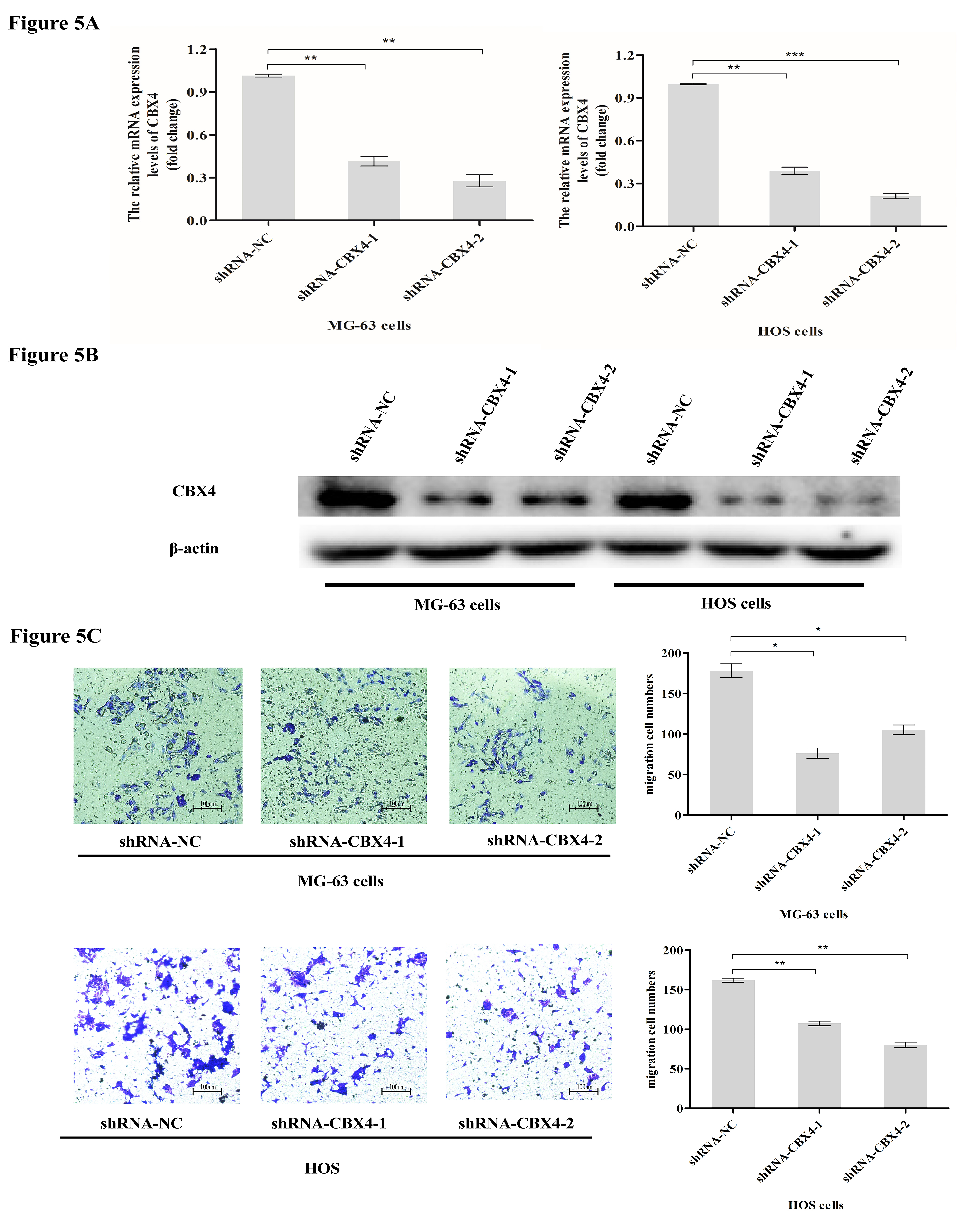 Fig. 5.
Fig. 5.CBX4 deficiency inhibits the metastasis of osteosarcoma cells.
(A,B) Lentiviral transduction was used to interfere with CBX4 expression in HOS
cells and MG-63 cells. (C) The migration ability was determined using the
indicated stable cells as described in the Methods section. The results showed
that silencing CBX4 significantly inhibited the metastasis of
osteosarcoma cells (HOS cells and MG-63 cells). The results are expressed as the
mean
METTL3 inhibits the metastasis of osteosarcoma cells through CBX4. To further demonstrate that CBX4 is involved in METTL3-mediated inhibition of osteosarcoma cell metastasis, we overexpressed CBX4 in HOS cells and MG-63 cells with METTL3 silencing. The results showed that the metastatic ability of osteosarcoma cells was attenuated after silencing METTL3 and enhanced after overexpression of CBX4, while there was no significant change in the metastatic ability of cells with overexpression of NC after silencing METTL3 (Fig. 6A,B). Negative Control (NC).
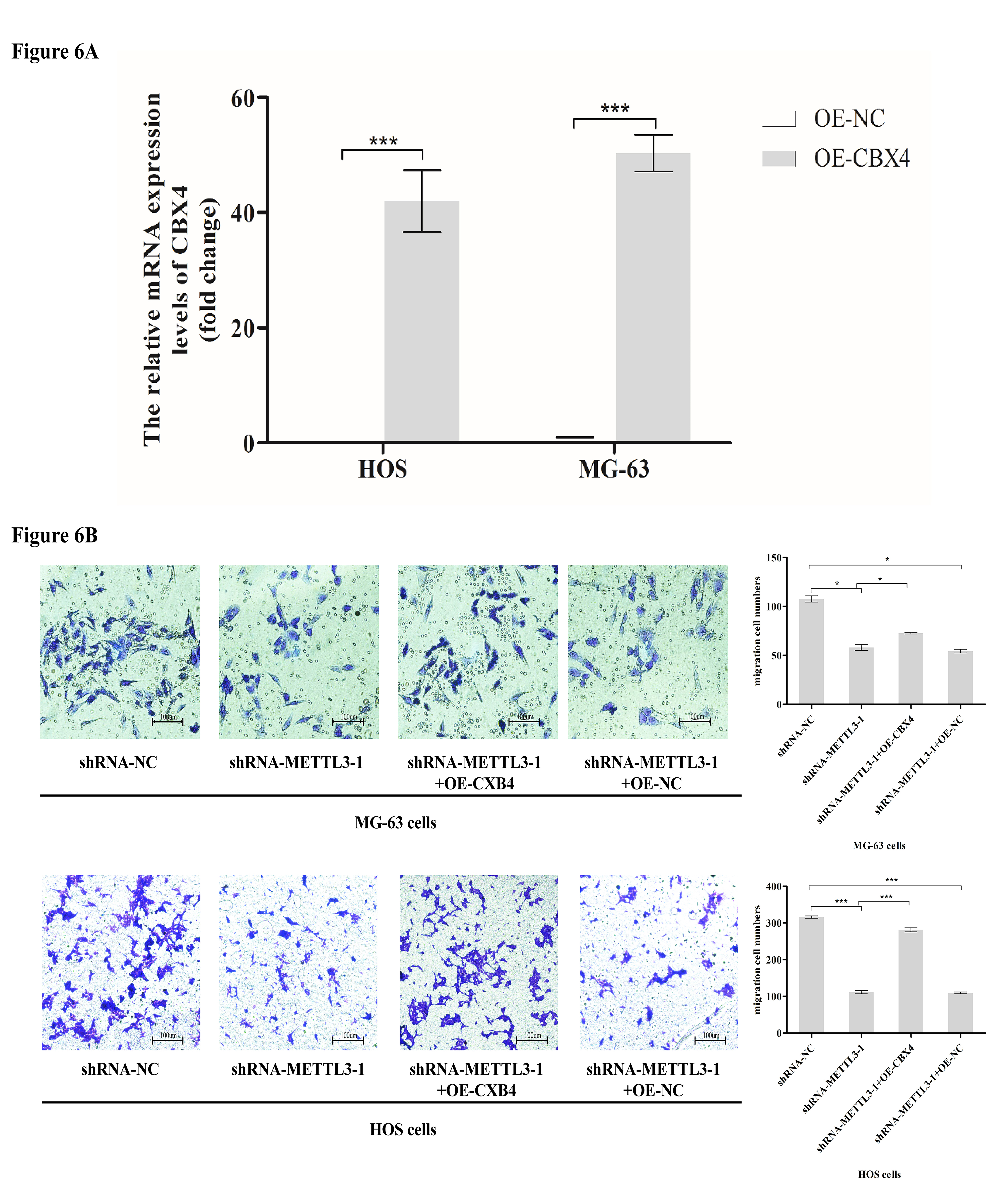 Fig. 6.
Fig. 6.METTL3 inhibits the metastasis of osteosarcoma cells through
CBX4. (A) Lentiviral transduction was used to overexpress CBX4 in HOS cells and
MG-63 cells. (B) The migration ability was determined using four groups of cells
(HOS cells and MG-63 cells): shRNA-NC, shRNA-METTL3-1, shRNA-METTL3-1+OE-CBX4,
and shRNA-METTL3-1+OE-NC. The results showed that the migration ability of
osteosarcoma cells was attenuated after silencing METTL3 and increased after
overexpression of CBX4, while there was no significant change in the metastatic
ability of cells with overexpression of NC after silencing METTL3. The results
are expressed as the mean
The specific regulatory mechanism of METTL3 in osteosarcoma is currently unknown. In this study, we used Western blotting to measure the levels of MMP-associated proteins (MMP2 and MMP9) and epithelial-mesenchymal transition (EMT)-associated proteins (E-Cadherin and N-Cadherin). The results showed that silencing METTL3 significantly decreased the protein expression of MMP2, MMP9, and N-Cadherin and increased the protein expression of E-Cadherin in osteosarcoma cells; moreover, overexpression of CBX4 increased the protein expression of MMP2, MMP9 and N-Cadherin and decreased the protein expression of E-Cadherin in osteosarcoma cells, and silencing METTL3 did not result in significant changes in these protein levels in cells with overexpression of NC (Fig. 7A,B).
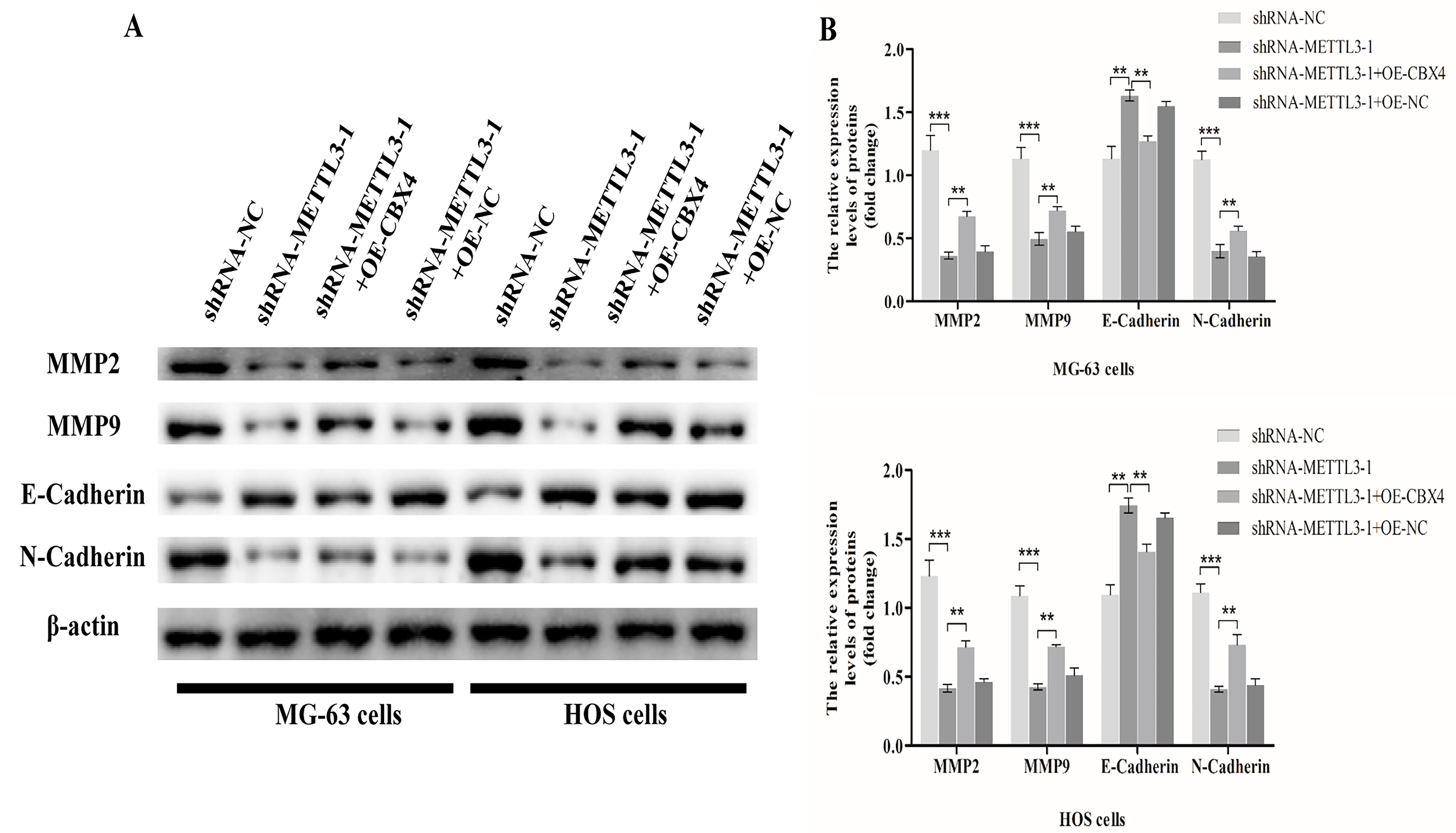 Fig. 7.
Fig. 7.METTL3 participates in the metastasis of osteosarcoma cells by
regulating matrix metalloproteinase (MMP) and epithelial-mesenchymal transition
(EMT) signaling through CBX4. (A) We used Western blotting to measure the levels
of MMP-associated proteins (MMP2 and MMP9) and EMT-associated proteins
(E-Cadherin and N-Cadherin) in four groups of cells (HOS cells and MG-63 cells):
shRNA-NC, shRNA-METTL3-1, shRNA-METTL3-1+OE-CBX4, and shRNA-METTL3-1+OE-NC. The
differences in protein (MMP2, MMP9, E-cadherin, N-cadherin) expression in the
four groups of cells were analyzed. The results are expressed as the mean
In previous studies, CBX4 was found to be involved in the invasion and metastasis of osteosarcoma cells and in epithelial-mesenchymal transition (EMT) [19, 22], while alterations in METTL3 expression promoted osteosarcoma progression [23, 24]. In this study, we investigated the molecular mechanism of METTL3 mediated through CBX4 in osteosarcoma metastasis.
METTL3, an important member of the m6A family, has been extensively investigated for its role in osteosarcoma. METTL3 has been found to be critically involved in tumor development in various cancers, including osteosarcoma [17]. Several studies have shown that METTL3 is highly expressed in sarcomas, and high levels of METTL3 are associated with poor prognosis [25, 26], consistent with the relationship between METTL3 expression and survival identified in our study by analysis of GEPIA data and whole-genome sequencing data from GSE12865. Numerous studies have shown that METTL3 is associated with tumor proliferation and metastasis [27, 28, 29]. To investigate the specific effects of METTL3 on OS cell metastasis and proliferation, we interfered with METTL3 expression. The results showed that METTL3 silencing inhibited the metastasis and proliferation of OS cells and promoted the apoptosis of OS cells. Taken together, these results suggest that METTL3 is highly expressed in osteosarcoma and is associated with poor prognosis and that silencing METTL3 inhibits the metastasis and proliferation of osteosarcoma cells and promotes their apoptosis.
CBX4 is a member of the CBX protein family and has been found to act as both a
tumor suppressor and an oncogene in cancer studies [30, 31]. Abnormal expression
of CBX4 is associated with tumorigenesis and cancer progression. In clear cell
renal cell carcinoma, CBX4 plays an oncogenic role by interacting with HDAC1 to
inhibit KLF6 transcription, CBX4 suppresses metastasis via recruitment of HDAC3
to the Runx2 gene promoter in colorectal carcinoma, and CBX4 is
upregulated and exerts a protumor effect in OS via activation of the
HIF-1
MMP2 and MMP9 are members of the matrix metalloproteinase (MMP) family that are essential for tumor metastasis by regulating invasion, angiogenesis, apoptosis, inflammation, cell growth, and the metastatic niche [35, 36, 37]. Studies have shown that HOXD11 promotes cell invasion and metastasis by activating the FN1/MMP2/MMP9 pathway in penile squamous cell carcinoma [38] and that UUSP19 enhances MMP2/MMP9-mediated tumorigenesis in gastric cancer [39]. We found that inhibition of METTL3 expression in osteosarcoma cells decreased the expression of MMP2 and MMP9, while overexpression of CBX4 restored their expression. We investigated the protein levels of both E-cadherin and N-Cadherin, which mediate critical EMT-related pathways to promote cancer cell invasion and metastasis [40]. Silencing METTL3 suppressed N-Cadherin expression and promoted E-Cadherin expression in osteosarcoma cells, while these changes in expression were reversed upon overexpression of CBX4. In summary, these results show that METTL3 regulates the expression of metastasis-associated genes in osteosarcoma cells through CBX4.
In summary, we elucidated the role of the METTL3-CBX4 axis in the regulation of osteosarcoma cell metastasis in this study, but there are still many precise molecular mechanisms to be investigated, such as the effect of METTL3 on osteosarcoma cell metastasis in vivo in nude mice and the pathway controlling the subsequent m6A methylation of CBX4 by METTL3, that is, which m6A reader is bound to the mRNA of CBX4. In the future, we will also investigate the role of CBX4 regulation by METTL3 on proliferation, apoptosis, EMT and other functions in osteosarcoma cells, aiming for the future treatment of osteosarcoma metastasis by combined intervention with METTL3 and CBX4 expression to improve the therapeutic effect in osteosarcoma.
The data and materials used in the current study are all available from the corresponding author upon reasonable request.
XWW and FSS made substantial contributions to the design of the paper; XSH, DL, DGC and MY made substantial contributions to the acquisition, analysis or interpretation of the working data; XSH and DL jointly analyzed the data and co-authored the manuscript; XWW and FSS reviewed and edited and finally approved the version to be published; All authors jointly were responsible for all aspects of the work and ensured that questions related to the accuracy or integrity of any part of the work were appropriately investigated and resolved. All authors read and approved the final manuscript. All authors contributed to editorial changes in the manuscript.
Not applicable.
The authors thank all students and technicians in the laboratory for their cooperation.
This study was funded by the National Natural Science Foundation of China (81703967).
Xi-song Huo as an employee of Shanghai Shuan’an Biotechnology Co. Itd, and Fu-sheng Shang the general manager of Shanghai Shuan’an Biotechnology Co. Itd. The authors declare no conflict of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
