- Academic Editor
-
-
-
Genome-wide association studies (GWAS) have mapped over 90% of disease- and
quantitative-trait-associated variants within the non-coding genome. Non-coding
regulatory DNA (e.g., promoters and enhancers) and RNA (e.g., 5
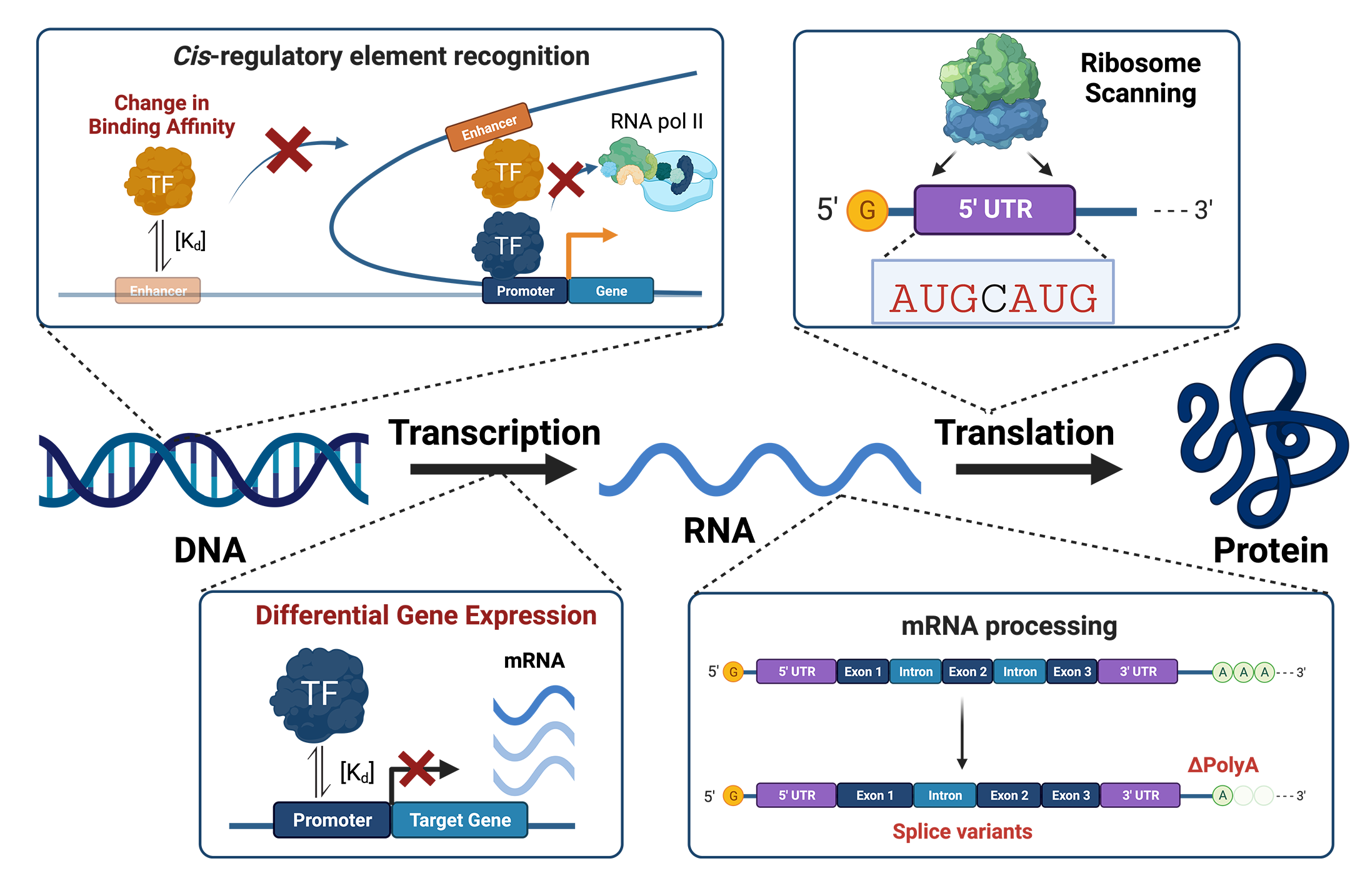
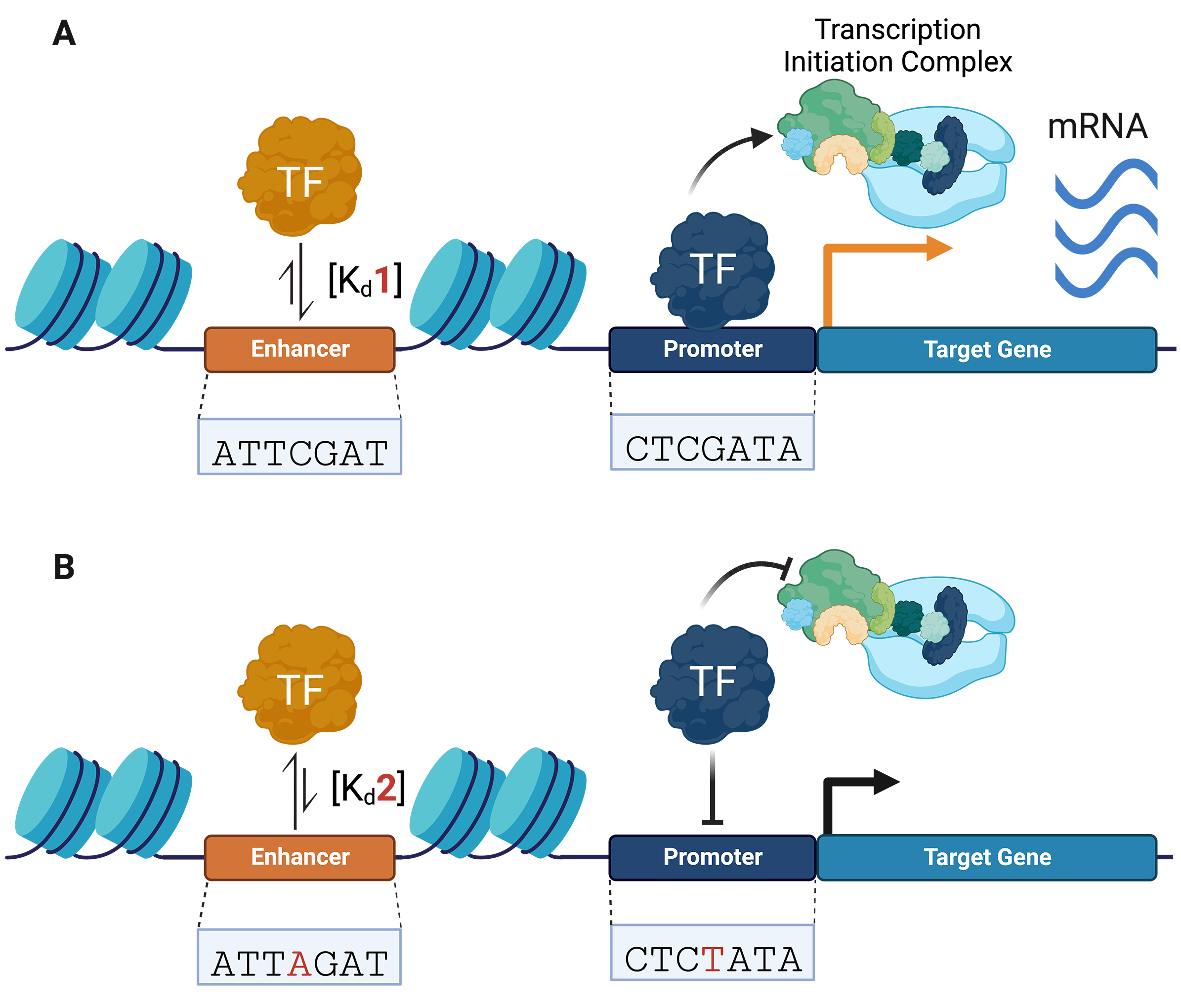
Genome-wide association studies (GWAS) have mapped over 90% of disease- and
quantitative-trait-associated variants within the non-coding genome. Non-coding
regulatory DNA (e.g., promoters and enhancers) and RNA (e.g., 5

