We show that down-regulation of circular ribonucleic acid 0001588 with small interfering-circular ribonucleic acid 0001588 mimics promoted caspase-3 and caspase-9 activity levels, increased lactate dehydrogenase activity levels, and reduced cell growth of in vitro model. Circular ribonucleic acid 0001588 plasmids increased circular ribonucleic acid 0001588 expressions and promoted cell growth and reduced activity levels of lactate dehydrogenase, caspase-3, and caspase-9 activity levels in vitro model of Alzheimer’s disease. Then, circular ribonucleic acid 0001588 down-regulation also promoted reactive oxygen species production and oxidative stress (malonaldehyde), and reduced superoxide dismutase, glutathione, and glutathione peroxidase levels by suppressing of silent information regulator 1/nuclear factor erythroid 2-related factor 2/heme oxygenase-1 in vitro. However, over-expression of circular ribonucleic acid 0001588 reduced reactive oxygen species production and malonaldehyde levels and increased superoxide dismutase, glutathione, and levels via activation signal pathway of silent information regulator 1/nuclear factor erythroid 2-related factor 2/heme oxygenase-1 signaling pathway by miR-211-5p up-regulation in vitro. Over-expression of miR-211-5p attenuated the role of circular ribonucleic acid 0001588 on Alzheimer’s disease-induced oxidative stress. Also, activation of the silent information regulator 1 pathway attenuated the antioxidative effects of circular ribonucleic acid 0001588 down-regulation on oxidative stress by activating silent information regulator 1/nuclear factor erythroid 2-related factor 2/heme oxygenase-1 in vitro. In conclusion, our findings indicated that circular ribonucleic acid 0001588 induced the silent information regulator 1/nuclear factor erythroid 2-related factor 2-dependent heme oxygenase-1 pathway to prevent oxidative stress by miR-211-5p in the rat model or in vitro model of a sporadic type of Alzheimer’s disease. Therefore, the expression of the circular ribonucleic acid 0001588 gene was inhibited in rodent Alzheimer’s disease model.
Alzheimer’s disease (AD) is a severe central neurodegeneration disorder related to aging, which has become one of the leading death causes following tumor and angiocardiopathy (Magro et al., 2015). AD is the most common cognition-related senile disease affecting the elderly (Li et al., 2013). AD is clinically manifested by gradually severe cognitive disorder, accompanied by irreversible behavior and human communication disorders. AD deterioration will deprive the patients of the ability of independent living (Cheng et al., 2016). Moreover, it will cause complications and even death with disease aggravation (Li et al., 2013). The pathological characteristics of AD mainly include extracellular amyloid protein (Aβ) deposition and intracellular neurofibrillary tangle (Cheng et al., 2016). However, the pathogenesis of AD remains unclear, involving multiple pathological changes, such as inflammatory response, oxidative stress, mitochondrial dysfunction, apoptosis, and synaptic dysfunction (Cheng et al., 2016; Reima et al., 2016).
Nuclear factor erythroid 2-related factor 2 (Nrf2) is a canonical nuclear transcription divisor regulating anti-oxidative stress response (Hong et al., 2014). Nrf2 activation can induce the expression of peroxiredoxin and enhance anti-oxidative stress ability. The function and role of Nrf2 in antioxidative stress has been intensively studied (Hong et al., 2014). Silent information regulator 1 (SIRT1) has been found to delay the pathogenesis and development of AD through anti-oxidative stress (Ak et al., 2015; Hsieh et al., 2009). Also, it plays a vital character in the elimination of AD pathological deposits, anti-inflammation, anti-apoptosis, and neurotrophy (Hsieh et al., 2009). In recent years, multiple single-target anti-AD agents have failed in clinical trials. Therefore, SIRT1/Nrf2 has been proposed as a possible important factor in achieving multi-target therapy of AD.
Circular ribonucleic acids (circRNAs) is a new class of functional molecule, which is 5’ to 3’-phosphodiester bond and consisting of a circular configuration (Wang et al., 2015). For more than 30 years, circRNAs expressed at differentially cancerous tissues and cell lines. CircRNAs have been reported to play crucial roles in the etiopathogenesis and development of various human kinds of diseases and acknowledged as products of gene rearrangement, aberrant splicing, or non-linear reverse splicing (Wang et al., 2015). MicroRNAs (miRNAs) are a variety of intrinsic endogenous non-coding small RNA molecules and target one or more mRNAs by binding with the 3’-untranslated region (3’-UTR) of target mRNAs (Kenny et al., 2018; Long et al., 2017). miRNAs can act against ischemia at the cellular level and can enhance adenosine triphosphate (ATP) synthesis and protect against free radical injury (Kenny et al., 2018; Long et al., 2017). Multiple studies have validated that miRNAs can prevent AD (Kenny et al., 2018). CircRNA 0000950 in AD has been reported to promote cell growth, suppress neurite outgrowth, and promote neural inflammatory cytokine levels via miRNA-103. Also, circular HDAC9/miRNA-138 pathway has been demonstrated to regulate synaptic and amyloid precursor protein processing deficits in AD (Browne et al., 2010; Yang et al., 2019).
The animals were 5-6-week age adult male Wistar rats maintained in a climate-controlled room (23 °C, 55-60 humidness) on an 8 : 00-20 : 00 (12 h/12 h light/dark cycle), provided with freedom chow diet and water ad libitum. All rats were randomly assigned into two groups: sham control group (n = 6) and AD model group (n = 6). Experimental Animal Ethical Committee of First Hospital of Shijiazhuang, P. R. China, had approved this study (No: 20160805-134234).
All rats of AD were anesthetized using pentobarbital sodium (Sangon Biotech, Shanghai, P. R. China) at a dose of 35 mg/kg, followed by exposure of skulls. Stainless steel gauge guide cannulae were inserted into the lateral ventricles. Stereotaxic coordinates were accurately located at 0.8 mm to bregma, at 1.5 mm to the sagittal suture, 3.6 mm to the surface of the brain. Rats were well-recovered after surgery and further subjected to 3 mg/kg of Streptozotocin on days 1 and 3 using a Hamilton syringe. Sham surgery animals were used as a control group. After 16 weeks, rats were anesthetized using pentobarbital sodium and sacrificed.
The left hemispheres were washed with PBS and fixed in 4% paraformaldehyde-phosphate buffer (0.1 M, pH 7.2) for 24 hr at room temperature. Tissue was embedded into the paraffin and cut into 4 μM-thick slices. Samples were stained with hematoxylin and eosin (H&E) and photographed under a light microscope (TE2000-U Nikon Eclipse, Japan).
In brief, rats were subjected to Morris water maze (Murakami et al., 2005; Zhao et al., 2005) twice/daily for five days. Rats were then placed on a probe trial on the sixth day and further placed on a visible platform trial on the seventh day. Water with milk power (30 ± 2 °C) was filled into a circular water tank (diameter, 120 cm; height, 60 cm). Afterward, rats were placed into the pool from different quadrants for training for 120 sec, and it was recorded. A rat was replaced on the platform for 20 sec, and the next training was performed after 120 sec of rest. After training for five days, the platform was removed, and the number of platform-crossings of rats within 120 sec was recorded.
Total RNA of serum samples was isolated using TRIzol reagent (Invitrogen, USA). CircRNA and miRNA were labeled using miRCURY™ Hy3™/Hy5™ Power labeling kit (Exiqon, Vedbaek, Denmark) and were hybridized using the miRCURYTM LNA Array (v.18.0) (Exiqon, Vedbaek, Denmark). Slides were scanned using the Axon GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA, USA).
Total RNA was extracted using TRIzol reagent (Invitrogen, USA). Then, total RNA was used to compound cDNA using reverse transcription (PrimeScript RT Regent Kit, Takara, Dalian, P. R. China). Q-PCR was performed using the SYBR PrimeScript RT-PCR Kit (Takara, Dalian, P. R. China). The amplification conditions were as follows: 95 °C for 10 min, 40 cycles of 95 °C for 30 sec, at 60 °C for 30 sec, and 72 °C for 30 sec.
Neuro-2a cell was cultured in Dulbecco’s Modified Eagle Medium (DMEM, Life Technologies, USA) containing 1% glutamine and 10% heat-inactivated fetal bovine serum (FBS, Life Technologies, USA) at 37 °C and 5% CO2. Neuro-2a cell was transfected with circRNA 0001588, anti-CircRNA 0001588, si-Nrf2, si-HO-1, and negative mimics using Lipofectamine® 2000 Reagent (Life Technologies). Neuro-2a cell was cultured and treated with Aβ (1-42) (10 µM, Life Technologies) for 24 hrs after transfection at 48 hrs.
Total sample proteins were isolated from the hippocampus by lysing in RIPA buffer (Beyotime) for 15 min at 4 °C and quantified using BCA assay (Beyotime). Afterward, 10 μg protein sample was used to measure the levels of malonaldehyde (MDA), superoxide dismutase (SOD), glutathione (GSH) and glutathione peroxidase (GSH-PX) using ELISA kits. Absorbance was measured at 450 nm in the SpectraMax M5 microplate reader (Molecular Devices Corp., Sunnyvale, CA, USA). The cell was stained using DCFH-DA (Beyotime, ROS Assay Kit) and observed using Zeiss Axioplan 2 (Carl Zeiss MicroImaging).
Lactate dehydrogenase (LDH) activity and caspase-3/9 activity levels were measured using LDH and caspase-3/caspase-9 activity kits (Beyotime). The absorbance of LDH activity or caspase-3/9 activity levels were measured using the SpectraMax M5 microplate reader (Molecular Devices Corp., Sunnyvale, CA, USA) at 450 nm or 405 nm.
Total protein was isolated from the hippocampus by lysing RIPA buffer (Beyotime) for 15 min at 4 °C and quantified using BCA protein assay (Beyotime). Every sample protein was subjected to 8-12% polyacrylamide gels, transferred onto polyvinylidene difluoride membranes (PVDF, GE Healthcare, Chalfont, UK). After blocking for 1 hr with 5%-non-milk in TBST at 37 °C, PVDF membranes were incubated with anti-SIRT1 (1 : 500, sc-135792, Santa Cruz Biotechnology, USA), anti-Nrf2 (1 : 500, sc-13032, Santa Cruz Biotechnology, USA), anti-HO-1 (1 : 500, Santa Cruz Biotechnology, USA) and anti-GAPDH (1 : 5000, Santa Cruz Biotechnology, Inc., Dallas, TX, USA) at 4 °C overnight. TBST was used to wash all membranes for 15 min, and membranes were incubated with anti-goat IgG secondary antibody at room temperature for 2 hrs. All membrane was visualized with Amersham ECL Western Blotting Detection Reagents (GE Healthcare, Amersham, Buckinghamshire, UK), and all protein blank was observed under LI-COR Odyssey gel imaging scanner (LI-COR Biosciences, Lincoln, NE, USA).
The cell was washed with PBS and fixed with 4% paraformaldehyde for 20 min. Cell blocked with 5% BSA containing 0.3% 0.1% Triton X-100 at 25 °C for 1 h and incubated with SIRT1 (1 : 100, Abcam; USA) at 4 °C overnight. The cell was washed with PBS for 15 min and incubated with Alexa Fluor 488 conjugated with anti-rabbit immunoglobulin G (IgG) (1 : 500 dilution) for 1 h. The cell was washed with PBS for 15 min and stained with DAPI for 15 min at darkness. The cell was captured using the Eclipse TE200 fluorescent microscope (Nikon, Melville, NY).
Data were presented as mean ± S.D. P < 0.05 was considered as statistical significance. All experiment data were analyzed using Student’s t-test for two groups or one-way analysis of variance (ANOVA) and Tukey’s post-test for multiple groups ( ≥ three groups).
Firstly, we analyzed the functions of circRNA 0001588 in a rat model of AD. As shown in Fig. 1A and 1B, the escape latency and the mean path length were significantly increased in the AD model group, compared with that in the control group. The time in the target quadrant and the number of times crossing the former platform location of AD rats were remarkably lower than those of control rats (Fig. 1C and 1D). Also, the MDA level was increased, while the levels of SOD, GSH, and GSH-px were reduced in AD model rats in comparison with the sham control group (Fig. 1E and 1H). HE (Hematoxylin-Eosin) staining revealed that the number of neurons was reduced at the CA1 area of the hippocampus in the AD model in comparison with the sham control group (Fig. 1I). Moreover, the expression of circRNA 0001588 was reduced in the AD model than that of the sham control group (Fig. 1K).
 Figure 1.
Figure 1.The expression of circRNA 0001588 in the AD model. The escape latency (A), the mean path length (B), The time in the target quadrant (C) and the number of times the animals crossed the former platform location (D), SOD (E), GSH (F), GSH-px (G), MDA (H), EH staining for neuro cell (I), heat map (J) and QPCR (K) for expression of circRNA 0001588 in AD model. **P < 0.01 versus the control group. Control, control group (n = 6); AD, AD model group (n = 6).
CircRNA 0001588 reduces oxidative stress in the AD rat model. In this study, we elucidated the effects of circRNA 0001588 pathway in oxidative stress in the AD rat model. CircRNA 0001588 mimics was used to increase the expression level of circRNA 0001588 (Fig. 2A). Over-expression of circRNA 0001588 promoted cell growth, while decreased the levels of LDH and caspase-3/9 activity in vitro of AD (Fig. 2B-2E). As shown in Fig. 2F-2K, the levels of MDA and ROS were decreased, while those of SOD, GSH, and GSH-px were increased following over-expression of circRNA 0001588 in vitro of the AD model, in comparison with the negative group. Next, anti-circRNA 0001588 mimics was utilized to decrease the expression levels of circRNA 0001588 in vitro of the AD model in contrast with the negative group (Fig. 3A). The down-regulation of circRNA 0001588 further reduced cell growth, and increased LDH and caspase-3/9 activity levels in vitro, in comparison with the negative group (Fig. 3B-3E). Also, the down-regulation of circRNA 0001588 increased the levels of MDA and ROS and decreased those of SOD, GSH, and GSH-px in vitro of the AD model (Fig. 3F-3K).
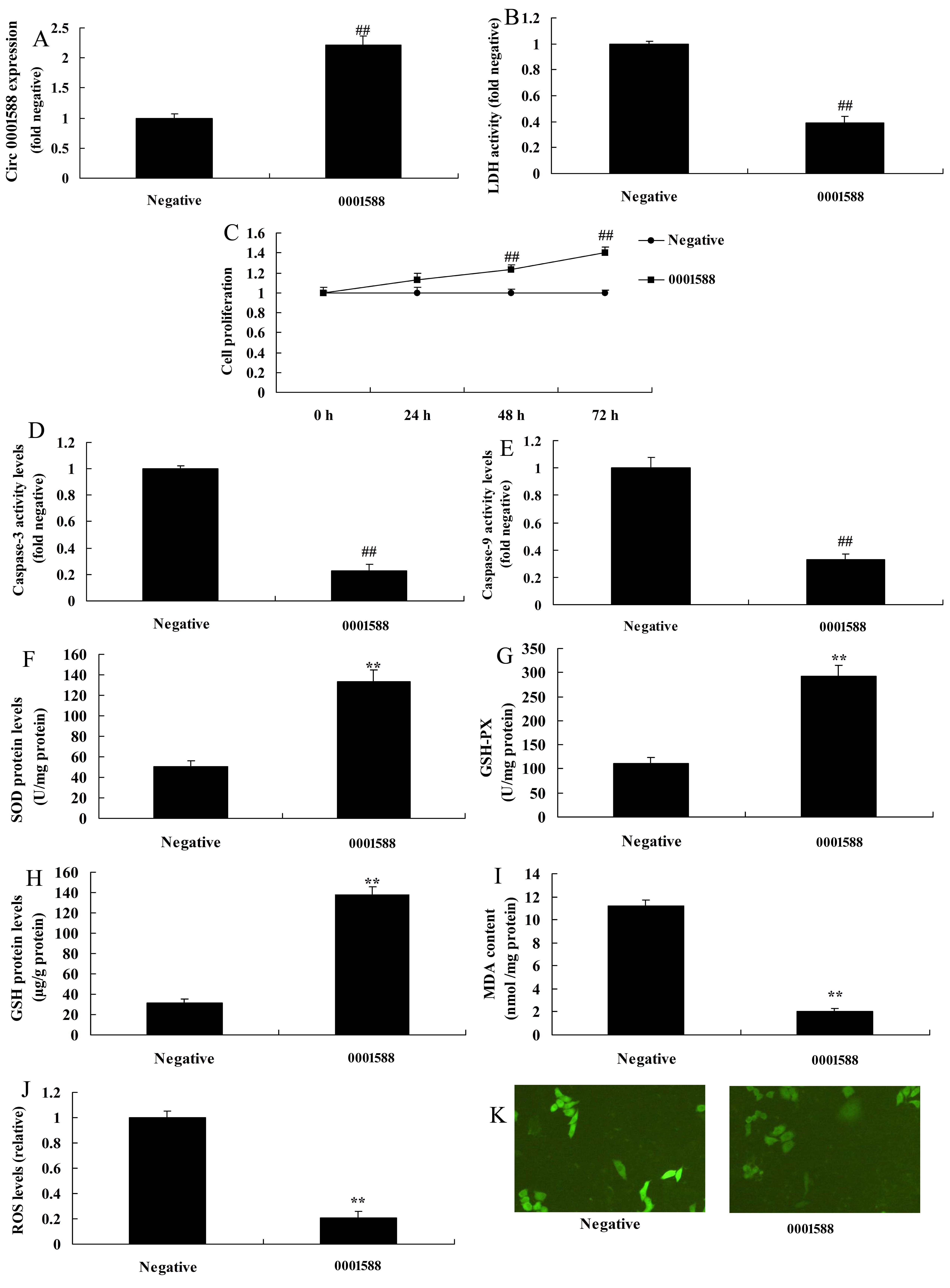 Figure 2.
Figure 2.CircRNA 0001588 regulates oxidative stress in a rat model of AD. QPCR (A) for expression of circRNA 0001588, LDH (B), cell growth (C), caspase-3/9 activity levels (D and E), SOD (F), GSH (G), GSH-px (H), MDA (I), and ROS (J and K) levels. **P < 0.01 versus the negative control group. Negative, negative control group (n = 3); circRNA 0001588, over-expression of circRNA 0001588 group (n = 3).
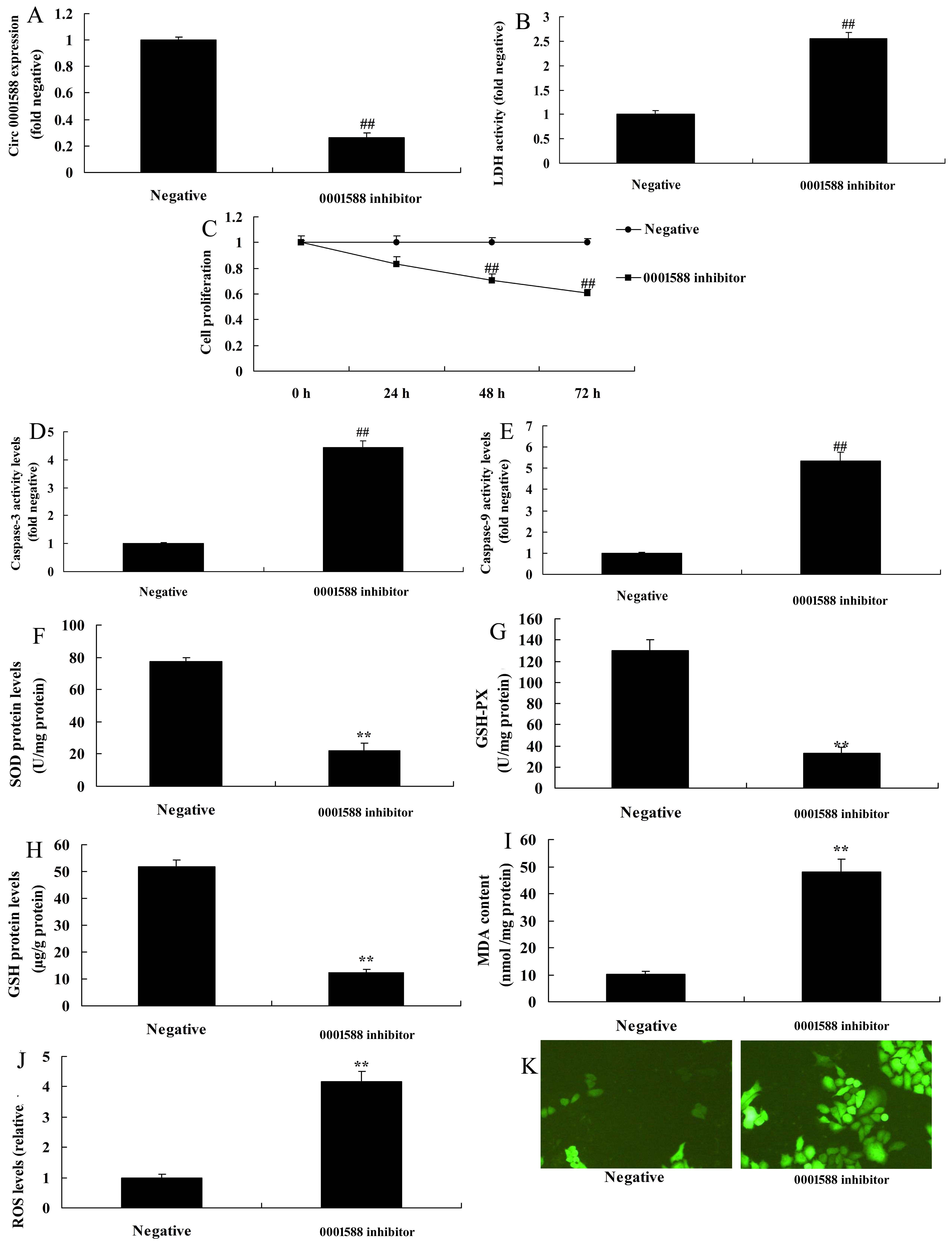 Figure 3.
Figure 3.Down-regulation of circRNA 0001588 regulates oxidative stress in a rat model of AD. QPCR (A) for expression of circRNA 0001588, LDH (B), cell growth (C), caspase-3/9 activity levels (D and E), SOD (F), GSH (G), GSH-px (H), MDA (I), and ROS (J and K) levels. **P < 0.01 versus the negative control group. Negative, negative control group (n = 3); anti-223-3p, down-regulation of circRNA 0001588 group (n = 3).
The study explained the mechanism of circRNA 0001588 in the AD rat model. Serum miR-211-5p expression levels were increased in a rat model of AD, comparing to the control group (Fig. 4A). Then, circRNA 0001588 was used to lessen the expression level of miR-211-5p in vitro, comparing to the negative group (Fig. 4B). The expression of circRNA 0001588 and miR-211-5p was negatively correlated (Fig. 4C). Luciferase activity assay revealed that circRNA 0001588 regulated the expression of miR-211-5p by directly targeting its mRNA 3’-UTR, and luciferase activity assay was reduced in circRNA 0001588 over-expression group (Fig. 4D and 4E). Also, the expression level of miR-211-5p was decreased in the circRNA 0001588 over-expression group, comparing to the negative group (Fig. 4F). However, the down-regulation of circRNA 0001588 increased the expression level of the miR-211-5p in vitro model (Fig. 4G).
 Figure 4.
Figure 4.CircRNA 0001588 regulates miR-211-5p in a rat model of AD. Gene chip for miR-211-5p in vivo model (A) and in vitro model (B), There was also a negative correlation between circRNA 0001588 and miR-211-5p (C), circRNA 0001588 regulates the expression of miR-211-5p by directly targeting its mRNA 3’-UTR (D), luciferase assay activity (E), miR-211-5p mRNA expression (F and G). Negative, negative control group (n = 3); anti-223-3p, down-regulation of circRNA 0001588 group (n = 3); miRNA-223-3p, over-expression of circRNA 0001588 group (n = 3). **P < 0.01 versus the negative control group.
Then, this study further determined the neuroprotective effects of circRNA 0001588 on ROS-induced oxidative stress in an AD rat model. The up-regulation of miR-211-5p suppressed the expression of Nrf2 and HO-1 protein in vitro model in comparison with the negative group (Fig. 5A). Luciferase activity assay revealed that miR-211-5p regulated the expression of the SIRT1 gene by directly targeting its mRNA 3’-UTR, and luciferase activity assay was weakened in the miR-211-5p group, comparing to negative group (Fig. 5B and 5C). IF assay showed that miR-211-5p up-regulation weakened the protein expression levels of SIRT1 in vitro, comparing to the negative group (Fig. 5D). Then, the up-regulation of miR-211-5p suppressed the protein expression levels of SIRT1, Nrf2, and HO-1 in vitro, in comparison with the negative group (Fig. 5E-5H). Next, circRNA 0001588 was administered to induce the protein expression levels of SIRT1, Nrf2, and HO-1 in vitro, compared with the negative group (Fig. 5I-5L).
 Figure 5.
Figure 5.CircRNA 0001588 regulates SIRT1/Nrf2/ HO-1 pathway in a rat model of AD by miR-211-5p. Gene chip for Nrf2/HO-1 pathway (A), circRNA 0001588 regulates the expression of SIRT1 by directly targeting its mRNA 3’-UTR (B), luciferase assay activity (C), SIRT1 protein expression by IF (D), SIRT1, Nrf2 and HO-1 protein expression using statistical analysis (E, F, and G), and western blotting analysis (H) by over-expression of circRNA 0001588; SIRT1, Nrf2 and HO-1 protein expression using statistical analysis (I, J and K), and western blotting analysis (L) by down-regulation of circRNA 0001588. **P < 0.01 versus the negative control group. Negative, negative control group (n = 3); anti-223-3p, down-regulation of circRNA 0001588 group (n = 3); miRNA-223-3p, over-expression of circRNA 0001588 group (n = 3).
To further study the function and mechanism of miR-211-5p in the effects of circRNA 0001588 on AD-induced oxidative stress, miR-211-5p was used to attenuate the effects of circRNA 0001588 on inhibiting miR-211-5p expression, activating the protein expression levels of SIRT1, Nrf2 and HO-1 in vitro, compared with circRNA 0001588 group (Fig. 6A-6E). The inhibition of Nrf2 pathway suppressed the effects of circRNA 0001588 on AD-induced the inhibition of cell growth, activation of LDH activity levels and caspase-3/9 activity levels, promotion of MDA and ROS levels, and inhibition of SOD, GSH and GSH-PX levels in vitro model, in comparison with circRNA 0001588 group (Fig. 6F-6O).
 Figure 6.
Figure 6.The miR-211-5p inhibited the effects of circRNA 0001588 on AD-induced oxidative stress. MiR-211-5p expression (A), SIRT1, Nrf2 and HO-1 protein expression using statistical analysis (B, C, and D), and western blotting analysis (E), LDH (F), cell growth (G), caspase-3/9 activity levels (H and I), SOD (J), GSH (K), GSH-px (L), MDA (M) and ROS (N and O) levels. **P < 0.01 versus negative control group, ##P < 0.01 versus negative control group. Negative, negative control group (n = 3); miRNA-223-3p, over-expression of circRNA 0001588 group (n = 3); si-Nrf2, miRNA-223-3p, over-expression of circRNA 0001588 and si-Nrf2 group (n = 3).
To further explore the function of SIRT1 in the effects of circRNA 0001588 on AD-induced oxidative stress, SIRT1 (20 μM of SIRT1 agonist, CAY10602) was employed to trigger the effects of circRNA 0001588 on the induction of SIRT1, Nrf2 and HO-1 protein expression in vitro (Fig. 7A-7D). Consequently, the induction of SIRT1 pathway suppressed the effects of anti-circRNA 0001588 on AD-induced the inhibition of cell growth, activation of LDH activity level and caspase-3/9 activity levels, promotion of MDA and ROS levels, and the inhibition levels of SOD, GSH and GSH-PX in vitro of AD model (Fig. 7E-7N).
 Figure 7.
Figure 7.The pathway inhibited the effects of anti-circRNA 0001588 on AD-induced oxidative stress. SIRT1, Nrf2 and HO-1 protein expression using statistical analysis (A, B and C), and western blotting analysis (D), LDH (E), cell growth (F), caspase-3/9 activity levels (G and H), SOD (I), GSH (J), GSH-px (K), MDA (L) and ROS (M and N) levels. **P < 0.01 versus negative control group, ##P < 0.01 versus negative control group. Negative, negative control group (n = 3); miRNA-223-3p, over-expression of circRNA 0001588 group (n = 3); si-Nrf2, miRNA-223-3p, over-expression of circRNA 0001588 and si-Nrf2 group (n = 3).
At present, the pathogenesis of AD remains unclear. There are multiple hypotheses, including the cholinergic hypothesis, the Aβ hypothesis, the Tau protein hypothesis, and the inflammation hypothesis (Cheng et al., 2016). Also, multiple pathophysiological changes can be observed in the brains of AD patients (Cheng et al., 2016), including senile plaque, neurofibrillary tangle, oxidative stress response, inflammatory response, and apoptosis (Kureel et al., 2014). There are complex correlations among these pathophysiological responses, which remain mostly undefined (Kureel et al., 2014; Zhang et al., 2016).
Our work revealed the expression levels of CircRNA 0001588 were down-regulated in the AD model, down-regulation of circRNA 0001588 increased the levels of MDA and ROS levels and reduced those levels of SOD, GSH, and GSH-px in vitro. Meanwhile, circRNA 0001588 down-regulation increased the expression level of miR-211-5p. MiR-211 gene regulates age-related cataracts that affected the antioxidant function of lens epithelial cells affected (Kim et al., 2015). These results showed that circRNA 0001588/miR-211-5p might regulated oxidative stress to adjust nerve cell death in the AD model.
The etiology of AD is exceptionally complicated (Panda et al., 2012). SIRT1 was first identified and named in 1994 (see Panda et al., 2012). Ever since then, a variety of studies have been performed to explore its role in antioxidative stress in all systems and organs (Panda et al., 2012). The role of SIRT1 in AD has been intensively studied. To be specific, SIRT1 has been demonstrated to exert effects on AD through antioxidative stress.
Moreover, recent studies have also found that SIRT1 can prevent or delay AD through multiple pathways (Browne et al., 2010; Zhang et al., 2015). Sun et al. (2019) have shown that dihydromyricetin exerts protective roles in AD by up-regulating the AMPK/SIRT1 pathway. Liu et al. (2018) have demonstrated that serum miR-211 expression levels, as a novel biomarker with high sensitivity and specificity, regulated SIRT1 expression, were affiliated to the pathogenesis and progression of diabetic retinopathy. Nrf2 can bind with Keap1 in the cytoplasm under physiological condition, however, in non-active and degradable status. In the presence of internal and external free radical and chemical stimulation, Nrf2 would dissociate with Keap1 (Augestad et al., 2014; Wu et al., 2017) to enter the nucleus and bind with antioxidant response element (ARE). Thus, Nrf2 initiates gene transcription to resist harmful internal and external stimulations, including the downstream two-phase detoxifying enzymes, peroxiredoxins, and proteasome/molecular chaperone of ARE (Wu et al., 2017). Our results have shown that the up-regulation of circRNA 0001588 in vitro of the AD model induced the protein expression levels of SIRT1, Nrf2, and HO-1 by miR-211-5p.
A recent study had indicated the existence of abnormal oxidative stress response in early AD, which is involved in the pathogenesis and progression of AD (Liu et al., 2017). Protein oxidation, lipid peroxidation, DNA, and RNA oxidation are remarkably enhanced in brain tissues of AD patients, transgenic AD mouse models, and the AD cell model (Liu et al., 2017). Also, activities of SOD, GSH, and CAT are reported to be decreased in brain tissues of AD patients (Gao et al., 2017). In APP and APP/PS1 transgenic mouse models, mRNA and protein levels of two-phase detoxifying enzyme NQO1, as well as GCL catalytic subunit (GCLC) and modifying subunit (GCLM) have been down-regulated, and NQO1 is decreased most significantly (Nagappan et al., 2017). Meanwhile, activities of SOD and glutathione peroxidase have also been down-regulated (Monzo et al., 2016). These results are consistent with the findings on human brain tissue (Monzo et al., 2016). These enzymes and antioxidants play vital roles in antioxidative stress response, and their expression is closely correlated with Nrf2 activation (Monzo et al., 2016). Therefore, enhancing the expression of downstream antioxidants targetingNrf2 can improve the antioxidative stress ability of the body. Moreover, Nrf2 plays a crucial role in the antioxidative stress response of AD (Nagappan et al., 2017). In the present model, the activation of the SIRT1 pathway suppressed the effects of circRNA 0001588 down-regulation on AD-induced oxidative stress. These indicated that circRNA 0001588 adversely targeted miR-211-5p, whereas miR-211-5p affects the SIRT1/Nrf2-dependent HO-1 pathway in AD.
In conclusion, we have shown that administration with circRNA 0001588 has notable benefits on neuroprotection function to inhibit the oxidative stress of AD by the induction of the SIRT1/Nrf2/HO-1 signal pathway by miR-211-5p. CircRNA 0001588/ miR-211-5p/SIRT1/Nrf2/Ho-1 pathway might be an effective therapeutic target for AD and which provides a novel therapeutic target for AD.
In conclusion, the activation of NF-κB and high expression of COX-2 in the brain of epileptic rats can be inhibited by κB-decoy and, in turn, inhibit inflammation and oxidative stress during epilepsy.
Experimental Animal Ethical Committee of First Hospital of Shijiazhuang, P. R. China, had approved this study (No: 20160805-134234).
The authors would also like to extend their thanks to reviewers for their opinions and suggestions.
The authors reported no potential conflict of interest.
