- Academic Editor
-
-
-
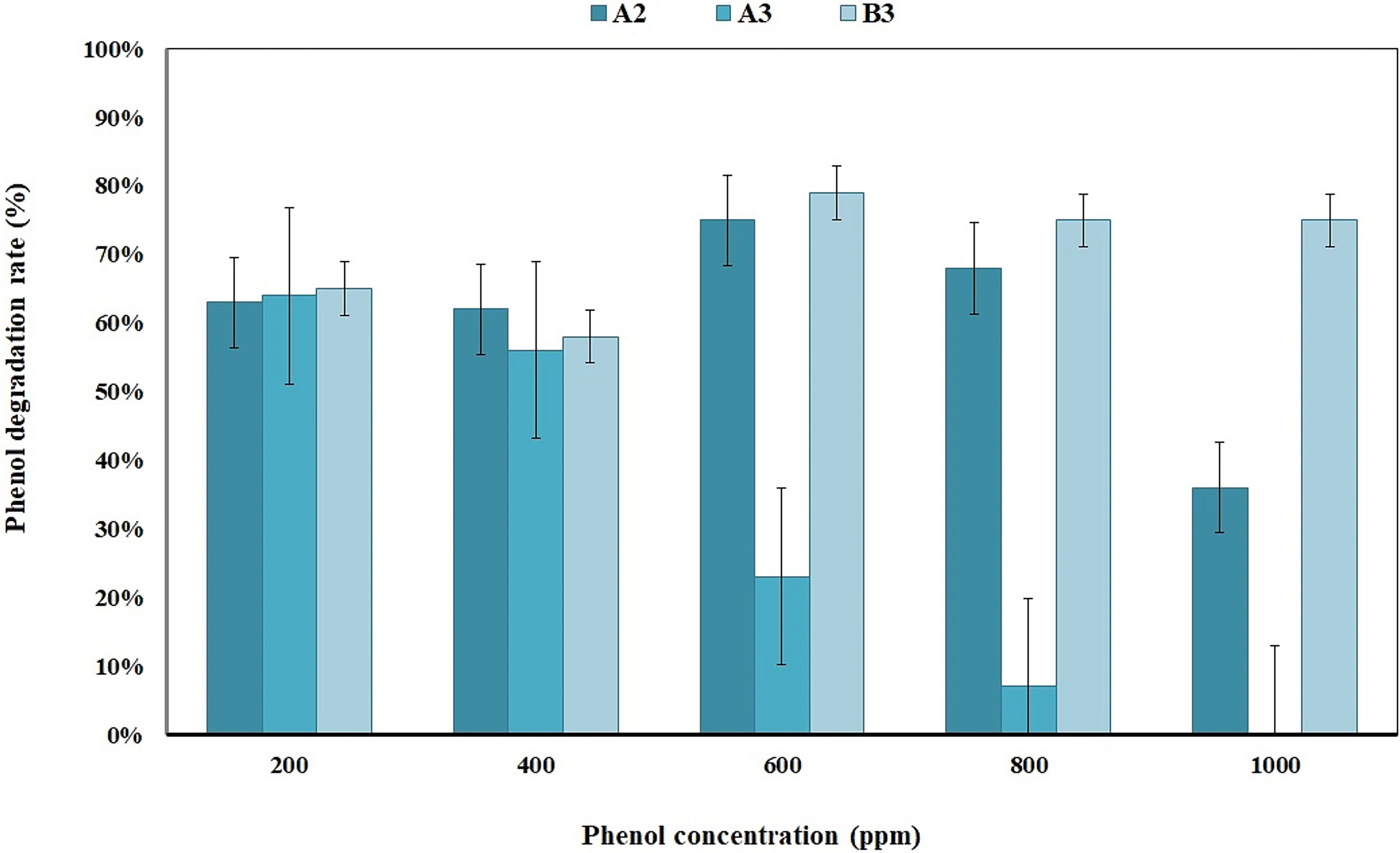
Background: By considering the importance and role of soil in the health of humanity, it is important to remove the presence of harmful compounds, such as phenol. Methods: In this study, four types of soil and leaf samples were collected from Kerman, Iran, and the amounts of heterotrophic and degradation bacteria were determined using the serial dilution and most probable number (MPN) methods. The amount of removed phenol was investigated using the Gibbs method with different concentrations of phenol. Then, an isolate with the highest percentage of phenol degradation was identified as the superior strain using 16 sRNA sequencing. The effects of the different factors, such as the carbon source (1% molasses and 1 g glucose), nitrogen source (0.1 g yeast extract), mixed culture, and time (14 and 28 days), on the biodegradation ability of the superior strain was investigated. Results: A total of 18 bacterial strains were isolated from the samples. Isolate B3 had the highest rate (75%) of phenol degradation, at a concentration of 1000 ppm, meaning it was identified as the superior strain. The molecular analysis results identified this isolate as the Comamonas testosteroni strain F4. This bacterium can degrade 89% of the phenol at 30 °C, 180 rpm, and 800 ppm over 28 days. C. testosteroni did not show a favorable phenol degradation ability in the presence of the investigated carbon sources, while this ability was also reduced in mixed cultures. Conclusions: C. testosteroni bacterial strain isolated from soil samples of pistachio orchards in Kerman, Iran, has a favorable ability to biodegrade phenol.