-
- Academic Editor
-
-
-
†These authors contributed equally.
Background: With the recent evolution of multidrug-resistant strains,
the genetic characteristics of foodborne Salmonella enterica serovar
Enteritidis and clinical isolates have changed. ST11 is now the most common
genotype associated with S. Enteritidis isolates. Methods: A
total of 83 strains of S. Enteritidis were collected at the General
Hospital of the People’s Liberation Army. Of these, 37 were from aseptic sites in
patients, 11 were from the feces of patients with diarrhea, and the remaining 35
were of chicken-origin. The minimum inhibitory concentration of S.
Enteritidis was determined by the broth microdilution method. Genomic DNA was
extracted using the QiAamp DNA Mini Kit, and whole-genome sequencing (WGS) was
performed using an Illumina X-ten platform. Prokka was used for gene prediction
and annotation, and bioinformatic analysis tools included Resfinder, ISFinder,
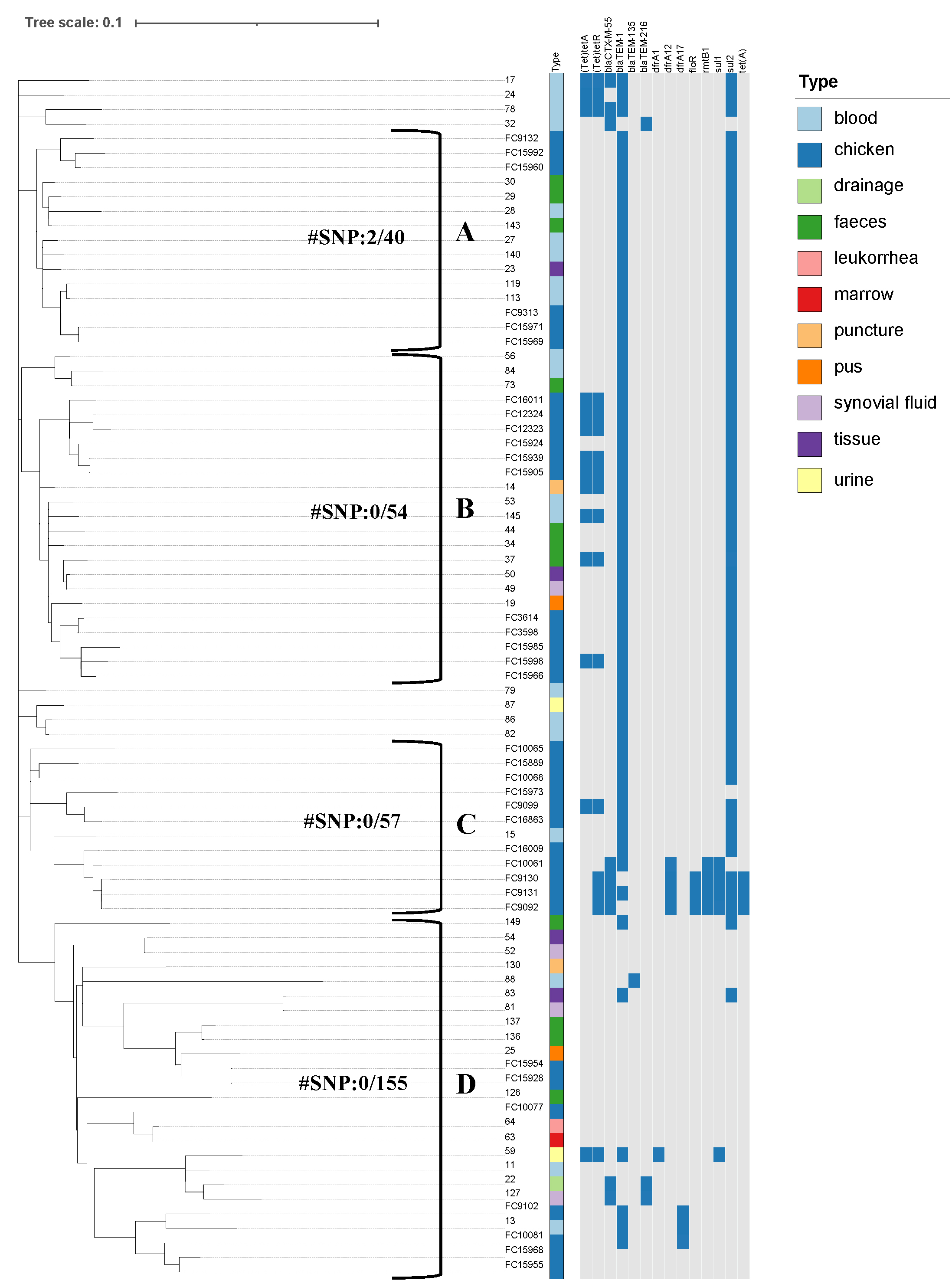
Virulence Factor Database, and PlasmidFinder. IQ-TREE was used to build a maximum
likelihood phylogenetic tree. The phylogenetic relationship and distribution of
resistance genes was displayed using iTOL. Comparative population genomics was
used to analyze the phenotypes and genetic characteristics of antibiotic
resistance in clinical and chicken-origin isolates of S. Enteritidis.
Results: The chicken-origin S. Enteritidis isolates were more
resistant to antibiotics than clinical isolates, and had a broader antibiotic
resistance spectrum and higher antibiotic resistance rate. A higher prevalence of
antibiotic-resistance genes was observed in chicken-origin S.
Enteritidis compared to clinical isolates, along with distinct patterns in the
contextual characteristics of these genes. Notably, genes such as
bla