-
- Academic Editor
-
-
-
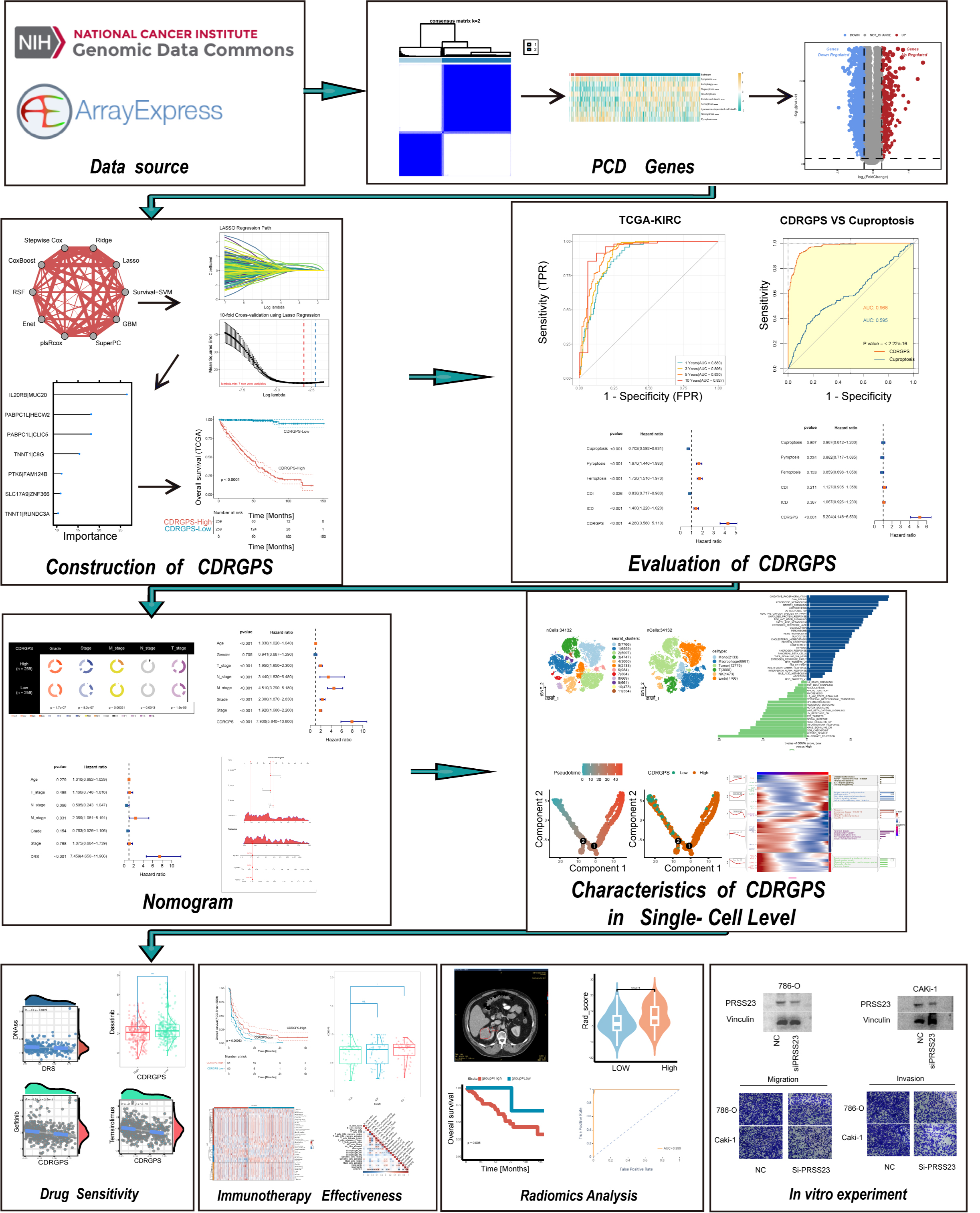
Background: Clear cell renal cell carcinoma (ccRCC) is a common and lethal urological malignancy for which there are no effective personalized therapeutic strategies. Programmed cell death (PCD) patterns have emerged as critical determinants of clinical prognosis and immunotherapy responses. However, the actual clinical relevance of PCD processes in ccRCC is still poorly understood. Methods: We screened for PCD-related gene pairs through single-sample gene set enrichment analysis (ssGSEA), consensus cluster analysis, and univariate Cox regression analysis. A novel machine learning framework incorporating 12 algorithms and 113 unique combinations were used to develop the cell death-related gene pair score (CDRGPS). Additionally, a radiomic score (Rad_Score) derived from computed tomography (CT) image features was used to classify the CDRGPS status as high or low. Finally, we conclusively verified the function of PRSS23 in ccRCC. Results: The CDRGPS was developed through an integrated machine learning approach that leveraged 113 algorithm combinations. CDRGPS represents an independent prognostic biomarker for overall survival and demonstrated consistent performance between training and external validation cohorts. Moreover, CDRGPS showed better prognostic accuracy compared to seven previously published cell death-related signatures. In addition, patients classified as high-risk by CDRGPS exhibited increased responsiveness to tyrosine kinase inhibitors (TKIs), mammalian Target of Rapamycin (mTOR) inhibitors, and immunotherapy. The Rad_Score demonstrated excellent discrimination for predicting high versus low CDRGPS status, with an area under the curve (AUC) value of 0.813 in the Cancer Imaging Archive (TCIA) database. PRSS23 was identified as a significant factor in the metastasis and immune response of ccRCC, thereby validating experimental in vitro results. Conclusions: CDRGPS is a robust and non-invasive tool that has the potential to improve clinical outcomes and enable personalized medicine in ccRCC patients.