-
- Academic Editors
-
-
-
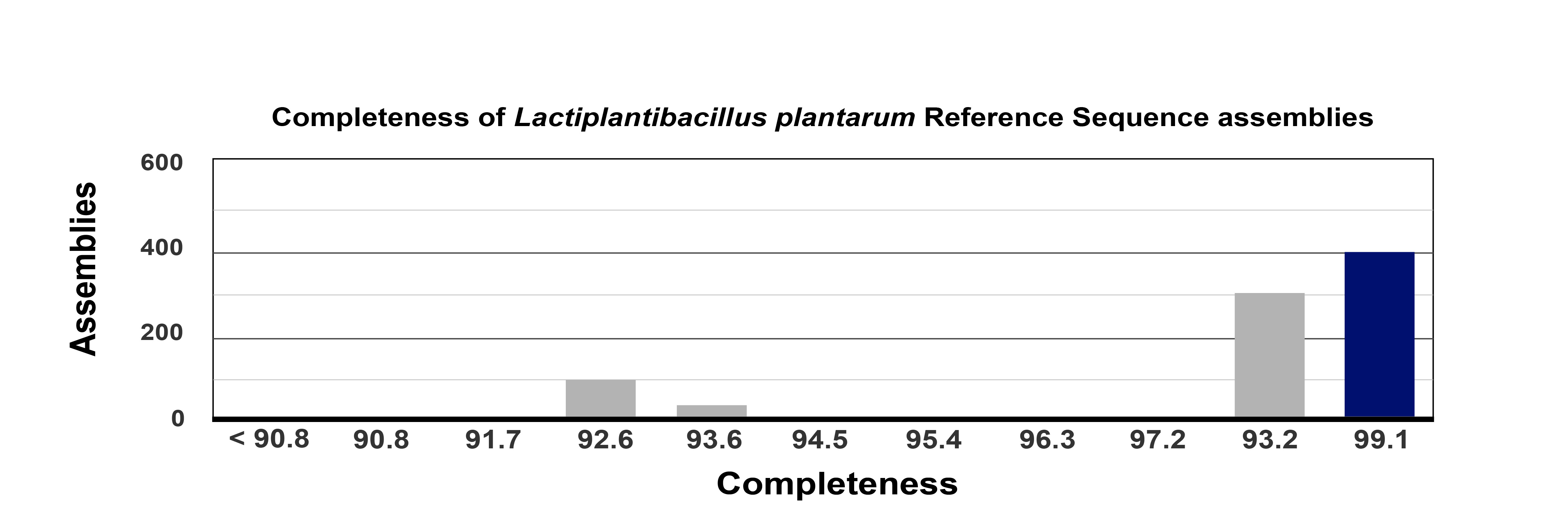
Background: Lactiplantibacillus plantarum 12-3 holds great promise as a probiotic bacterial strain, yet its full potential remains untapped. This study aimed to better understand this potential therapeutic strain by exploring its genomic landscape, genetic diversity, CRISPR-Cas mechanism, genotype, and mechanistic perspectives for probiotic functionality and safety applications. Methods: L. plantarum 12-3 was isolated from Tibetan kefir grains and, subsequently, Illumina and Single Molecule Real-Time (SMRT) technologies were used to extract and sequence genomic DNA from this organism. After performing pan-genomic and phylogenetic analysis, Average Nucleotide Identity (ANI) was used to confirm the taxonomic identity of the strain. Antibiotic resistance gene analysis was conducted using the Comprehensive Antibiotic Resistance Database (CARD). Antimicrobial susceptibility testing, and virulence gene identification were also included in our genomic analysis to evaluate food safety. Prophage, genomic islands, insertion sequences, and CRISPR-Cas sequence analyses were also carried out to gain insight into genetic components and defensive mechanisms within the bacterial genome. Results: The 3.4 Mb genome of L. plantarum 12-3, was assembled with 99.1% completeness and low contamination. A total of 3234 genes with normal length and intergenic spacing were found using gene prediction tools. Pan-genomic studies demonstrated gene diversity and provided functional annotation, whereas phylogenetic analysis verified taxonomic identity. Our food safety study revealed a profile of antibiotic resistance that is favorable for use as a probiotic. Analysis of insertional sequences, genomic islands, and prophage within the genome provided information regarding genetic components and their possible effects on evolution. Conclusions: Pivotal genetic elements uncovered in this study play a crucial role in bacterial defense mechanisms and offer intriguing prospects for future genome engineering efforts. Moreover, our findings suggest further in vitro and in vivo studies are warranted to validate the functional attributes and probiotic potential of L. plantarum 12-3. Expanding the scope of the research to encompass a broader range of L. plantarum 12-3 strains and comparative analyses with other probiotic species would enhance our understanding of this organism’s genetic diversity and functional properties.