-
- Academic Editor
-
-
-
†These authors contributed equally.
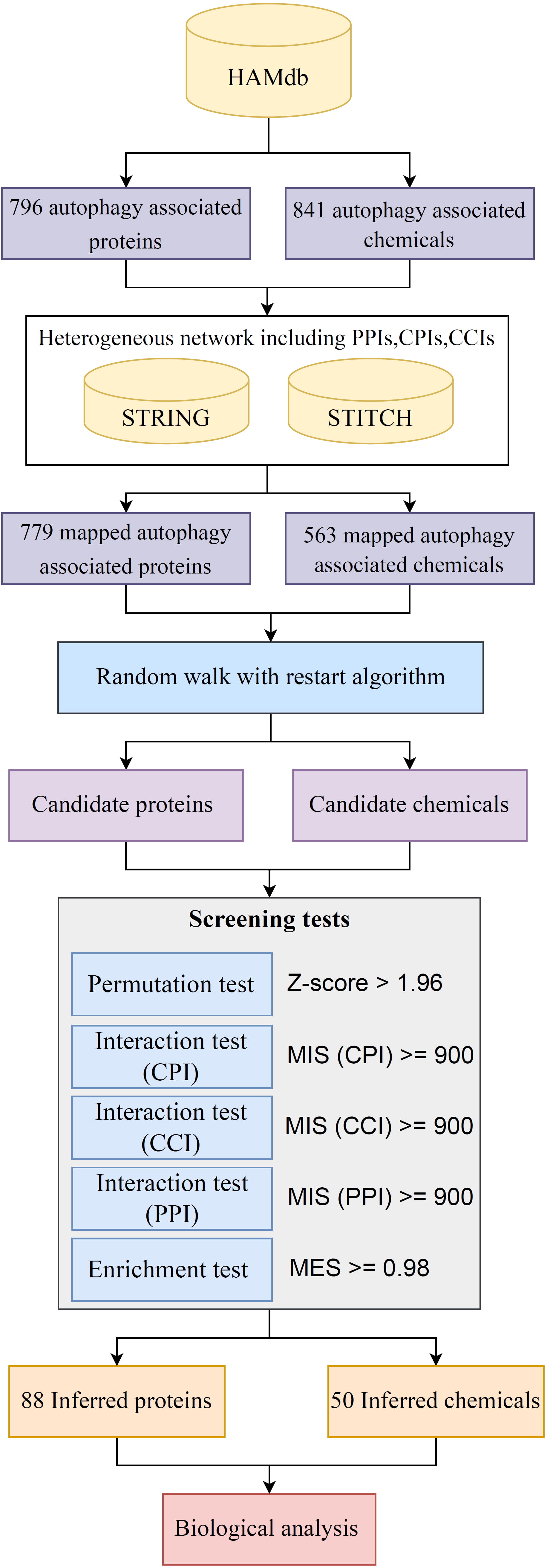
Background: Autophagy is instrumental in various health conditions, including cancer, aging, and infections. Therefore, examining proteins and compounds associated with autophagy is paramount to understanding cellular biology and the origins of diseases, paving the way for potential therapeutic and disease prediction strategies. However, the complexity of autophagy, its intersection with other cellular pathways, and the challenges in monitoring autophagic activity make the experimental identification of these elements arduous. Methods: In this study, autophagy-related proteins and chemicals were catalogued on the basis of Human Autophagy-dedicated Database. These entities were mapped to their respective PubChem identifications (IDs) for chemicals and Ensembl IDs for proteins, yielding 563 chemicals and 779 proteins. A network comprising protein–protein, protein–chemical, and chemical–chemical interactions was probed employing the Random-Walk-with-Restart algorithm using the aforementioned proteins and chemicals as seed nodes to unearth additional autophagy-associated proteins and chemicals. Screening tests were performed to exclude proteins and chemicals with minimal autophagy associations. Results: A total of 88 inferred proteins and 50 inferred chemicals of high autophagy relevance were identified. Certain entities, such as the chemical prostaglandin E2 (PGE2), which is recognized for modulating cell death-induced inflammatory responses during pathogen invasion, and the protein G Protein Subunit Alpha I1 (GNAI1), implicated in ether lipid metabolism influencing a range of cellular processes including autophagy, were associated with autophagy. Conclusions: The discovery of novel autophagy-associated proteins and chemicals is of vital importance because it enhances the understanding of autophagy, provides potential therapeutic targets, and fosters the development of innovative therapeutic strategies and interventions.